当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Late-Stage Reshaping of Phage-Displayed Libraries to Macrocyclic and Bicyclic Landscapes using a Multipurpose Linchpin
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2024-12-19 , DOI: 10.1021/jacs.4c13561 Kejia Yan, Mark Miskolzie, Fernando Banales Mejia, Chuanhao Peng, Arunika I. Ekanayake, Alexey Atrazhev, Jessica Cao, Dustin J. Maly, Ratmir Derda
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2024-12-19 , DOI: 10.1021/jacs.4c13561 Kejia Yan, Mark Miskolzie, Fernando Banales Mejia, Chuanhao Peng, Arunika I. Ekanayake, Alexey Atrazhev, Jessica Cao, Dustin J. Maly, Ratmir Derda
|
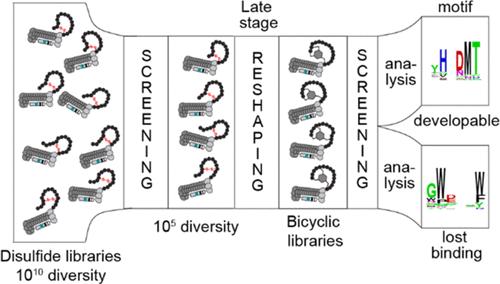
Genetically encoded libraries (GEL) are increasingly being used for the discovery of ligands for “undruggable” targets that cannot be addressed with small molecules. Foundational GEL platforms like phage-, yeast-, ribosome-, and mRNA-display have enabled the display of libraries composed of 20 natural amino acids (20AA). Unnatural amino acids (UAA) and chemical post-translational modification (cPTM) expanded GEL beyond the 20AA space to yield unnatural linear, cyclic, and bicyclic peptides. The standard operating procedure incorporates UAA and cPTM into a “naive” library with 108–1012 compounds and uses a chemically upgraded library in multiple rounds of selection to discover target-binding hits. However, such an approach uses zero knowledge of natural peptide-receptor interactions that might have been discovered in selections performed with 20AA libraries. There is currently no consensus regarding whether “zero-knowledge” naive libraries or libraries with pre-existing knowledge can offer a more effective path to discovery of molecular interactions. In this manuscript, we evaluated the feasibility of discovery of macrocyclic and bicyclic peptides from “nonzero-knowledge” libraries. We approach this problem by late-stage chemical reshaping of a preselected phage-displayed landscape of 20AA binders to NS3aH1 protease. The reshaping is performed using a novel multifunctional C2-symmetric linchpin, 3,5-bis(bromomethyl)benzaldehyde (termed KYL), that combines two electrophiles that react with thiols and an aldehyde group that reacts with N-terminal amine. KYL diversified phage-displayed peptides into bicyclic architectures and delineated 2 distinct sequence populations: (i) peptides with the HXDMT motif that retained binding upon bicyclization and (ii) peptides without the HXDMT motif that lost binding once chemically modified. The same HXDMT family can be found in traditional selections starting from the naive KYL-modified library. Our report provides a case study for discovering advanced, chemically upgraded macrocycles and bicycles from libraries with pre-existing knowledge. The results imply that other selection campaigns completed in 20AA space, potentially, can serve for late-stage reshaping and as a starting point for the discovery of advanced peptide-derived ligands.
更新日期:2024-12-20






























 京公网安备 11010802027423号
京公网安备 11010802027423号