Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Systematic characterization of indel variants using a yeast-based protein folding sensor
Structure ( IF 4.4 ) Pub Date : 2024-12-19 , DOI: 10.1016/j.str.2024.11.017 Sven Larsen-Ledet, Søren Lindemose, Aleksandra Panfilova, Sarah Gersing, Caroline H. Suhr, Aitana Victoria Genzor, Heleen Lanters, Sofie V. Nielsen, Kresten Lindorff-Larsen, Jakob R. Winther, Amelie Stein, Rasmus Hartmann-Petersen
Structure ( IF 4.4 ) Pub Date : 2024-12-19 , DOI: 10.1016/j.str.2024.11.017 Sven Larsen-Ledet, Søren Lindemose, Aleksandra Panfilova, Sarah Gersing, Caroline H. Suhr, Aitana Victoria Genzor, Heleen Lanters, Sofie V. Nielsen, Kresten Lindorff-Larsen, Jakob R. Winther, Amelie Stein, Rasmus Hartmann-Petersen
|
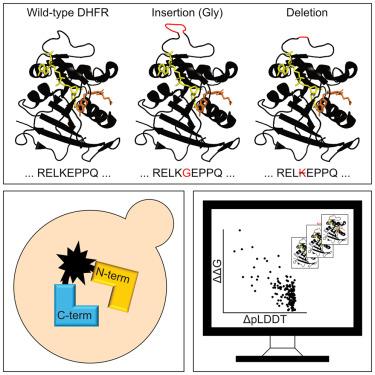
Gene variants resulting in insertions or deletions of amino acid residues (indels) have important consequences for evolution and are often linked to disease, yet, compared to missense variants, the effects of indels are poorly understood and predicted. We developed a sensitive protein folding sensor based on the complementation of uracil auxotrophy in yeast by circular permutated orotate phosphoribosyltransferase (CPOP). The sensor reports on the folding of disease-linked missense variants and de -novo -designed proteins. Applying the folding sensor to a saturated library of single-residue indels in human dihydrofolate reductase (DHFR) revealed that most regions that tolerate indels are confined to internal loops, the termini, and a central α helix. Several indels are temperature sensitive, and folding is rescued upon binding to methotrexate. Rosetta and AlphaFold2 predictions correlate with the observed effects, suggesting that most indels destabilize the native fold and that these computational tools are useful for the classification of indels observed in population sequencing.
中文翻译:

使用基于酵母的蛋白质折叠传感器系统表征插入缺失变体
导致氨基酸残基 (插入缺失) 插入或缺失的基因变异对进化有重要影响,并且通常与疾病有关,然而,与错义变异相比,插入缺失的影响知之甚少,预测能力不足。我们开发了一种灵敏的蛋白质折叠传感器,该传感器基于环状置换乳清酸磷酸核糖转移酶 (CPOP) 对酵母中尿嘧啶营养缺陷型的互补作用。该传感器报告了疾病相关的错义变体和从头设计的蛋白质的折叠。将折叠传感器应用于人二氢叶酸还原酶 (DHFR) 中饱和的单残基插入缺失文库,发现大多数耐受插入缺失的区域仅限于内环、末端和中央α螺旋。几个插入缺失对温度敏感,在与甲氨蝶呤结合时折叠被挽救。Rosetta 和 AlphaFold2 预测与观察到的效果相关,表明大多数插入缺失会破坏天然折叠的稳定性,并且这些计算工具对于在群体测序中观察到的插入缺失进行分类很有用。
更新日期:2024-12-19
中文翻译:

使用基于酵母的蛋白质折叠传感器系统表征插入缺失变体
导致氨基酸残基 (插入缺失) 插入或缺失的基因变异对进化有重要影响,并且通常与疾病有关,然而,与错义变异相比,插入缺失的影响知之甚少,预测能力不足。我们开发了一种灵敏的蛋白质折叠传感器,该传感器基于环状置换乳清酸磷酸核糖转移酶 (CPOP) 对酵母中尿嘧啶营养缺陷型的互补作用。该传感器报告了疾病相关的错义变体和从头设计的蛋白质的折叠。将折叠传感器应用于人二氢叶酸还原酶 (DHFR) 中饱和的单残基插入缺失文库,发现大多数耐受插入缺失的区域仅限于内环、末端和中央α螺旋。几个插入缺失对温度敏感,在与甲氨蝶呤结合时折叠被挽救。Rosetta 和 AlphaFold2 预测与观察到的效果相关,表明大多数插入缺失会破坏天然折叠的稳定性,并且这些计算工具对于在群体测序中观察到的插入缺失进行分类很有用。

































 京公网安备 11010802027423号
京公网安备 11010802027423号