当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Specific Response Assembly of 3D Space-Confined DNA Nanoaggregates for Rapid and Sensitive Detection of DNA Methyltransferase
Analytical Chemistry ( IF 6.7 ) Pub Date : 2024-12-18 , DOI: 10.1021/acs.analchem.4c05563 Jia-Hao Tang, Wei Liu, Mao-Hua Gao, Xian-Ming Guo, Yan-Mei Lei, Ying Zhuo
Analytical Chemistry ( IF 6.7 ) Pub Date : 2024-12-18 , DOI: 10.1021/acs.analchem.4c05563 Jia-Hao Tang, Wei Liu, Mao-Hua Gao, Xian-Ming Guo, Yan-Mei Lei, Ying Zhuo
|
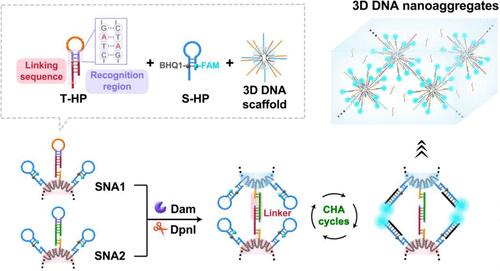
Rapid and sensitive detection of DNA adenine methyltransferase (Dam) activity is crucial for both research and clinical applications. Herein, we utilize two types of spherical nucleic acids (SNAs) to specific response assemble into 3D space-confined DNA nanoaggregates that enable the rapid and sensitive detection of Dam activity. The SNAs feature 3D order DNA scaffolds that serve as cores for anchoring signal hairpin probes (S-HPs) and target hairpin probes (T-HPs). Specifically, two distinct S-HPs are labeled with FAM fluorophores and BHQ1 quenchers and share identical hairpin sequences, while two types of T-HPs are designed with different linking sequences and specific recognition regions, resulting in the formation of two types of SNAs (SNA1 and SNA2). In the presence of Dam, the recognition region of the T-HPs is methylated and subsequently cleaved by auxiliary endonuclease, releasing the loop of the T-HP as a walking strand and exposing the linking sequence on the SNAs. Notably, the prior design of complementary linking sequences in the two types of SNAs facilitates their assembly into 3D DNA nanoaggregates, creating a confined space for walking strands to recover fluorescent signals. The 3D DNA nanoaggregate system not only provides highly ordered tracks but also enhances the spatial continuity of the walking strands, greatly improving the reaction kinetics for detecting Dam activity. This strategy enables the rapid and sensitive detection of Dam activity within 105 min, achieving a limit of detection of 2.9 × 10–4 U mL–1, demonstrating significant potential for advancing research in DNA methylation.
中文翻译:

用于快速、灵敏检测 DNA 甲基转移酶的 3D 空间限制 DNA 纳米聚集体的特异性响应组装
快速、灵敏地检测 DNA 腺嘌呤甲基转移酶 (Dam) 活性对于研究和临床应用都至关重要。在此,我们利用两种类型的球形核酸 (SNA) 对特异性响应组装成 3D 空间限制的 DNA 纳米聚集体,从而能够快速、灵敏地检测 Dam 活性。SNA 具有 3D 顺序 DNA 支架,用作锚定信号发夹探针 (S-HP) 和靶发夹探针 (T-HP) 的核心。具体来说,两种不同的 S-HP 用 FAM 荧光团和 BHQ1 淬灭基团标记,并共享相同的发夹序列,而两种类型的 T-HP 设计具有不同的连接序列和特异性识别区域,从而形成两种类型的 SNA(SNA1 和 SNA2)。在 Dam 存在下,T-HPs 的识别区被甲基化,随后被辅助核酸内切酶切割,将 T-HP 的环作为行走链释放出来,并暴露 SNA 上的连接序列。值得注意的是,两种 SNA 中互补连接序列的先前设计有助于它们组装成 3D DNA 纳米聚集体,为行走链创造一个狭窄的空间以恢复荧光信号。3D DNA 纳米聚集体系统不仅提供了高度有序的轨迹,还增强了行走链的空间连续性,大大改善了检测 Dam 活性的反应动力学。该策略能够在 105 分钟内快速、灵敏地检测 Dam 活性,实现 2.9 × 10–4 U mL–1 的检测限,显示出推进 DNA 甲基化研究的巨大潜力。
更新日期:2024-12-19
中文翻译:

用于快速、灵敏检测 DNA 甲基转移酶的 3D 空间限制 DNA 纳米聚集体的特异性响应组装
快速、灵敏地检测 DNA 腺嘌呤甲基转移酶 (Dam) 活性对于研究和临床应用都至关重要。在此,我们利用两种类型的球形核酸 (SNA) 对特异性响应组装成 3D 空间限制的 DNA 纳米聚集体,从而能够快速、灵敏地检测 Dam 活性。SNA 具有 3D 顺序 DNA 支架,用作锚定信号发夹探针 (S-HP) 和靶发夹探针 (T-HP) 的核心。具体来说,两种不同的 S-HP 用 FAM 荧光团和 BHQ1 淬灭基团标记,并共享相同的发夹序列,而两种类型的 T-HP 设计具有不同的连接序列和特异性识别区域,从而形成两种类型的 SNA(SNA1 和 SNA2)。在 Dam 存在下,T-HPs 的识别区被甲基化,随后被辅助核酸内切酶切割,将 T-HP 的环作为行走链释放出来,并暴露 SNA 上的连接序列。值得注意的是,两种 SNA 中互补连接序列的先前设计有助于它们组装成 3D DNA 纳米聚集体,为行走链创造一个狭窄的空间以恢复荧光信号。3D DNA 纳米聚集体系统不仅提供了高度有序的轨迹,还增强了行走链的空间连续性,大大改善了检测 Dam 活性的反应动力学。该策略能够在 105 分钟内快速、灵敏地检测 Dam 活性,实现 2.9 × 10–4 U mL–1 的检测限,显示出推进 DNA 甲基化研究的巨大潜力。






























 京公网安备 11010802027423号
京公网安备 11010802027423号