当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Unveiling the enzymatic pathway of UMG-SP2 urethanase: insights into polyurethane degradation at the atomic level
Chemical Science ( IF 7.6 ) Pub Date : 2024-12-18 , DOI: 10.1039/d4sc06688j P. Paiva, L. M. C. Teixeira, R. Wei, W. Liu, G. Weber, J. P. Morth, P. Westh, A. R. Petersen, M. B. Johansen, A. Sommerfeldt, A. Sandahl, D. E. Otzen, P. A. Fernandes, M. J. Ramos
Chemical Science ( IF 7.6 ) Pub Date : 2024-12-18 , DOI: 10.1039/d4sc06688j P. Paiva, L. M. C. Teixeira, R. Wei, W. Liu, G. Weber, J. P. Morth, P. Westh, A. R. Petersen, M. B. Johansen, A. Sommerfeldt, A. Sandahl, D. E. Otzen, P. A. Fernandes, M. J. Ramos
|
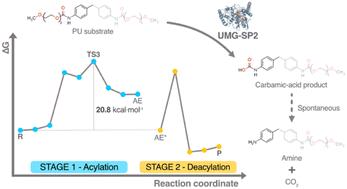
The recently discovered metagenomic urethanases UMG-SP1, UMG-SP2, and UMG-SP3 have emerged as promising tools to establish a bio-based recycling approach for polyurethane (PU) waste. These enzymes are capable of hydrolyzing urethane bonds in low molecular weight dicarbamates as well as in thermoplastic PU and the amide bond in polyamide employing a Ser-Sercis-Lys triad for catalysis, similar to members of the amidase signature protein superfamily. Understanding the catalytic mechanism of these urethanases is crucial for enhancing their enzymatic activity and improving PU bio-recycling processes. In this study, we employed hybrid quantum mechanics/molecular mechanics methods to delve into the catalytic machinery of the UMG-SP2 urethanase in breaking down a model PU substrate. Our results indicate that the reaction proceeds in two stages: STAGE 1 – acylation, in which the enzyme becomes covalently bound to the PU substrate, releasing an alcohol-leaving group; STAGE 2 – deacylation, in which a catalytic water hydrolyzes the enzyme:ligand covalent adduct, releasing the product in the form of a highly unstable carbamic acid, expected to rapidly decompose into an amine and carbon dioxide. We found that STAGE 1 comprises the rate-limiting step of the overall reaction, consisting of the cleavage of the substrate's urethane bond by its ester moiety and the release of the alcohol-leaving group (overall Gibbs activation energy of 20.8 kcal mol−1). Lastly, we identified point mutations that are expected to enhance the enzyme's turnover for the hydrolysis of urethane bonds by stabilizing the macrodipole of the rate-limiting transition state. These findings expand our current knowledge of urethanases and homolog enzymes from the amidase signature superfamily, paving the way for future research on improving the enzymatic depolymerization of PU plastic materials.
中文翻译:

揭示 UMG-SP2 氨基甲酸酶的酶途径:原子水平聚氨酯降解的见解
最近发现的宏基因组氨基甲酸酶 UMG-SP1、UMG-SP2 和 UMG-SP3 已成为建立聚氨酯 (PU) 废物生物基回收方法的有前途的工具。这些酶能够水解低分子量二氨基甲酸酯以及热塑性 PU 中的氨基甲酸酯键,以及聚酰胺中的酰胺键,采用 Ser-Ser顺式-Lys 三联体进行催化,类似于酰胺酶特征蛋白超家族的成员。了解这些氨基甲酸酶的催化机制对于增强其酶活性和改进 PU 生物回收过程至关重要。在这项研究中,我们采用混合量子力学/分子力学方法深入研究了 UMG-SP2 氨基甲酸乙酯酶在分解模型 PU 底物中的催化机制。我们的结果表明,反应分两个阶段进行:阶段 1 – 酰化,其中酶与 PU 底物共价结合,释放出离醇基团;第 2 阶段 – 脱酰基化,其中催化水解酶:配体共价加合物,以高度不稳定的氨基甲酸形式释放产物,预计会迅速分解成胺和二氧化碳。我们发现第 1 阶段包括整个反应的限速步骤,包括底物的酯部分裂解底物的氨基甲酸酯键和释放醇离开基团(总吉布斯活化能为 20.8 kcal mol-1)。最后,我们确定了有望通过稳定限速过渡态的大偶极子来增强酶对氨基甲酸酯键水解的转换的点突变。 这些发现扩展了我们目前对酰胺酶特征超家族的氨基甲酸酶和同系物酶的了解,为未来改善 PU 塑料材料酶解聚的研究铺平了道路。
更新日期:2024-12-18
中文翻译:

揭示 UMG-SP2 氨基甲酸酶的酶途径:原子水平聚氨酯降解的见解
最近发现的宏基因组氨基甲酸酶 UMG-SP1、UMG-SP2 和 UMG-SP3 已成为建立聚氨酯 (PU) 废物生物基回收方法的有前途的工具。这些酶能够水解低分子量二氨基甲酸酯以及热塑性 PU 中的氨基甲酸酯键,以及聚酰胺中的酰胺键,采用 Ser-Ser顺式-Lys 三联体进行催化,类似于酰胺酶特征蛋白超家族的成员。了解这些氨基甲酸酶的催化机制对于增强其酶活性和改进 PU 生物回收过程至关重要。在这项研究中,我们采用混合量子力学/分子力学方法深入研究了 UMG-SP2 氨基甲酸乙酯酶在分解模型 PU 底物中的催化机制。我们的结果表明,反应分两个阶段进行:阶段 1 – 酰化,其中酶与 PU 底物共价结合,释放出离醇基团;第 2 阶段 – 脱酰基化,其中催化水解酶:配体共价加合物,以高度不稳定的氨基甲酸形式释放产物,预计会迅速分解成胺和二氧化碳。我们发现第 1 阶段包括整个反应的限速步骤,包括底物的酯部分裂解底物的氨基甲酸酯键和释放醇离开基团(总吉布斯活化能为 20.8 kcal mol-1)。最后,我们确定了有望通过稳定限速过渡态的大偶极子来增强酶对氨基甲酸酯键水解的转换的点突变。 这些发现扩展了我们目前对酰胺酶特征超家族的氨基甲酸酶和同系物酶的了解,为未来改善 PU 塑料材料酶解聚的研究铺平了道路。

































 京公网安备 11010802027423号
京公网安备 11010802027423号