当前位置:
X-MOL 学术
›
Biotechnol. Bioeng.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Oleaginous Yeast Biology Elucidated With Comparative Transcriptomics
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2024-12-11 , DOI: 10.1002/bit.28891 Sarah J. Weintraub, Zekun Li, Carter L. Nakagawa, Joseph H. Collins, Eric M. Young
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2024-12-11 , DOI: 10.1002/bit.28891 Sarah J. Weintraub, Zekun Li, Carter L. Nakagawa, Joseph H. Collins, Eric M. Young
|
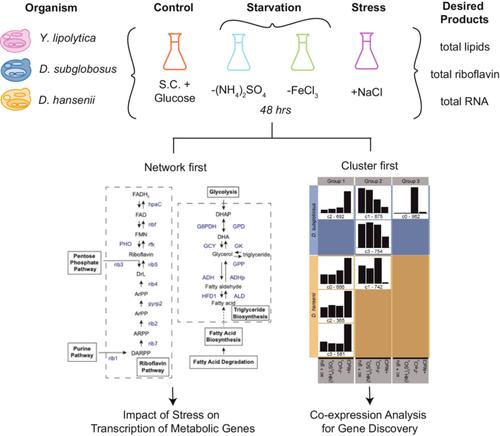
Extremophilic yeasts have favorable metabolic and tolerance traits for biomanufacturing‐ like lipid biosynthesis, flavinogenesis, and halotolerance – yet the connection between these favorable phenotypes and strain genotype is not well understood. To this end, this study compares the phenotypes and gene expression patterns of biotechnologically relevant yeasts Yarrowia lipolytica , Debaryomyces hansenii , and Debaryomyces subglobosus grown under nitrogen starvation, iron starvation, and salt stress. To analyze the large data set across species and conditions, two approaches were used: a “network‐first” approach where a generalized metabolic network serves as a scaffold for mapping genes and a “cluster‐first” approach where unsupervised machine learning co‐expression analysis clusters genes. Both approaches provide insight into strain behavior. The network‐first approach corroborates that Yarrowia upregulates lipid biosynthesis during nitrogen starvation and provides new evidence that riboflavin overproduction in Debaryomyces yeasts is overflow metabolism that is routed to flavin cofactor production under salt stress. The cluster‐first approach does not rely on annotation; therefore, the coexpression analysis can identify known and novel genes involved in stress responses, mainly transcription factors and transporters. Therefore, this work links the genotype to the phenotype of biotechnologically relevant yeasts and demonstrates the utility of complementary computational approaches to gain insight from transcriptomics data across species and conditions.
中文翻译:

用比较转录组学阐明的油质酵母生物学
极端嗜酸酵母菌对生物制造具有良好的代谢和耐受性特征,如脂质生物合成、黄素生成和耐盐性——但这些有利的表型与菌株基因型之间的联系尚不清楚。为此,本研究比较了在氮饥饿、铁饥饿和盐胁迫下生长的生物技术相关酵母 Yarrowia lipolytica、Debaryomyces hansenii 和 Debaryomyces subglobosus 的表型和基因表达模式。为了分析跨物种和条件的大型数据集,使用了两种方法:“网络优先”方法,其中广义代谢网络作为绘制基因的支架,以及“簇优先”方法,其中无监督机器学习共表达分析将基因聚集。这两种方法都提供了对应变行为的洞察。网络优先方法证实了 Yarrowia 在氮饥饿期间上调脂质生物合成,并提供了新的证据,证明 Debaryomyces 酵母中的核黄素过量生产是溢出代谢,在盐胁迫下被路由到黄素辅因子的产生。集群优先方法不依赖于 Annotation;因此,共表达分析可以鉴定参与应激反应的已知和新基因,主要是转录因子和转运蛋白。因此,这项工作将基因型与生物技术相关酵母的表型联系起来,并展示了互补计算方法的效用,以从不同物种和条件的转录组学数据中获得洞察力。
更新日期:2024-12-11
中文翻译:

用比较转录组学阐明的油质酵母生物学
极端嗜酸酵母菌对生物制造具有良好的代谢和耐受性特征,如脂质生物合成、黄素生成和耐盐性——但这些有利的表型与菌株基因型之间的联系尚不清楚。为此,本研究比较了在氮饥饿、铁饥饿和盐胁迫下生长的生物技术相关酵母 Yarrowia lipolytica、Debaryomyces hansenii 和 Debaryomyces subglobosus 的表型和基因表达模式。为了分析跨物种和条件的大型数据集,使用了两种方法:“网络优先”方法,其中广义代谢网络作为绘制基因的支架,以及“簇优先”方法,其中无监督机器学习共表达分析将基因聚集。这两种方法都提供了对应变行为的洞察。网络优先方法证实了 Yarrowia 在氮饥饿期间上调脂质生物合成,并提供了新的证据,证明 Debaryomyces 酵母中的核黄素过量生产是溢出代谢,在盐胁迫下被路由到黄素辅因子的产生。集群优先方法不依赖于 Annotation;因此,共表达分析可以鉴定参与应激反应的已知和新基因,主要是转录因子和转运蛋白。因此,这项工作将基因型与生物技术相关酵母的表型联系起来,并展示了互补计算方法的效用,以从不同物种和条件的转录组学数据中获得洞察力。






























 京公网安备 11010802027423号
京公网安备 11010802027423号