当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Computer Folding of Parallel DNA G‐Quadruplex: Hitchhiker's Guide to the Conformational Space
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-12-10 , DOI: 10.1002/jcc.27535 Michal Janeček, Petra Kührová, Vojtěch Mlýnský, Petr Stadlbauer, Michal Otyepka, Giovanni Bussi, Jiří Šponer, Pavel Banáš
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-12-10 , DOI: 10.1002/jcc.27535 Michal Janeček, Petra Kührová, Vojtěch Mlýnský, Petr Stadlbauer, Michal Otyepka, Giovanni Bussi, Jiří Šponer, Pavel Banáš
|
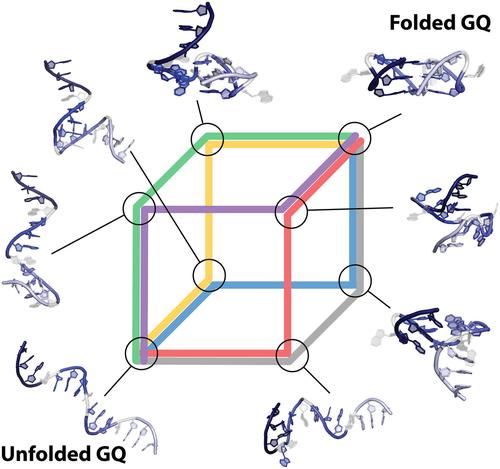
Guanine quadruplexes (GQs) play crucial roles in various biological processes, and understanding their folding pathways provides insight into their stability, dynamics, and functions. This knowledge aids in designing therapeutic strategies, as GQs are potential targets for anticancer drugs and other therapeutics. Although experimental and theoretical techniques have provided valuable insights into different stages of the GQ folding, the structural complexity of GQs poses significant challenges, and our understanding remains incomplete. This study introduces a novel computational protocol for folding an entire GQ from single‐strand conformation to its native state. By combining two complementary enhanced sampling techniques, we were able to model folding pathways, encompassing a diverse range of intermediates. Although our investigation of the GQ free energy surface (FES) is focused solely on the folding of the all‐anti parallel GQ topology, this protocol has the potential to be adapted for the folding of systems with more complex folding landscapes.
中文翻译:

平行 DNA G-四链体的计算机折叠:构象空间漫游指南
鸟嘌呤四链体 (GQ) 在各种生物过程中起着至关重要的作用,了解它们的折叠途径有助于深入了解它们的稳定性、动力学和功能。这些知识有助于设计治疗策略,因为 GQ 是抗癌药物和其他疗法的潜在靶点。尽管实验和理论技术为 GQ 折叠的不同阶段提供了有价值的见解,但 GQ 的结构复杂性带来了重大挑战,我们的理解仍然不完整。本研究引入了一种新的计算协议,用于将整个 GQ 从单链构象折叠到其天然状态。通过结合两种互补的增强采样技术,我们能够对折叠途径进行建模,包括各种中间体。尽管我们对 GQ 自由能表面 (FES) 的研究仅集中在全反并行 GQ 拓扑的折叠上,但该协议有可能适用于具有更复杂折叠景观的系统折叠。
更新日期:2024-12-10
中文翻译:

平行 DNA G-四链体的计算机折叠:构象空间漫游指南
鸟嘌呤四链体 (GQ) 在各种生物过程中起着至关重要的作用,了解它们的折叠途径有助于深入了解它们的稳定性、动力学和功能。这些知识有助于设计治疗策略,因为 GQ 是抗癌药物和其他疗法的潜在靶点。尽管实验和理论技术为 GQ 折叠的不同阶段提供了有价值的见解,但 GQ 的结构复杂性带来了重大挑战,我们的理解仍然不完整。本研究引入了一种新的计算协议,用于将整个 GQ 从单链构象折叠到其天然状态。通过结合两种互补的增强采样技术,我们能够对折叠途径进行建模,包括各种中间体。尽管我们对 GQ 自由能表面 (FES) 的研究仅集中在全反并行 GQ 拓扑的折叠上,但该协议有可能适用于具有更复杂折叠景观的系统折叠。































 京公网安备 11010802027423号
京公网安备 11010802027423号