Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Analysis of spider silk in loading-unloading cycles using Raman spectroscopy based on molecular bioinformatics of spidrion
Polymer ( IF 4.1 ) Pub Date : 2024-12-06 , DOI: 10.1016/j.polymer.2024.127910 Yi-qin Hong, Xin-ru Zhang, Li-Hua Wu, Tai-Yong Lv, Gustavo V. Guinea, José Pérez-Rigueiro, Ping Jiang
Polymer ( IF 4.1 ) Pub Date : 2024-12-06 , DOI: 10.1016/j.polymer.2024.127910 Yi-qin Hong, Xin-ru Zhang, Li-Hua Wu, Tai-Yong Lv, Gustavo V. Guinea, José Pérez-Rigueiro, Ping Jiang
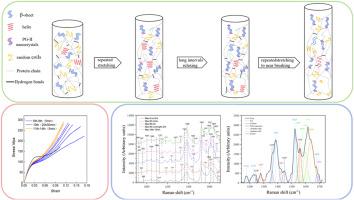
|
The mechanical properties of spider silk result from its organization at various levels, including the amino acid sequence, protein structure, protein assembly and full-hierarchical microstructure. However, the relatively few reports that contain an analysis of the motifs along the full-length of the sequences, and of the evolution of their secondary structure when the fiber is subjected to mechanical load, render difficult the task of relating sequence, microstructure and properties for this material. In this study, we identified seven spider major ampullate gland silk proteins of Argiope bruennichi , determine their full-length amino acid motifs, simulated the repeated stretching of spider major ampullate gland silk (MAS) in the natural environment, verified the stability of its mechanical properties, and established the evolution of its protein structure by semi-quantitative analysis of Raman spectroscopy. After stretching at different strains, MAS can recover previous mechanical behavior and exhibit excellent shape and mechanical memory in terms of longitudinal stretching. It is also shown that MAS maintain its mechanical properties through a precise adjustment of protein structure, secondary structure transformation and reconstruction. This study shows that the repeated stretching characteristics of spider major ampullate gland silk may be a post-processing adjustment way that, in combination with its molecular organization, may bring a new inspiration for the relation of sequence-structure-property.
中文翻译:

使用基于 spidrion 分子生物信息学的拉曼光谱分析装载-卸载循环中的蜘蛛丝
蜘蛛丝的机械性能是其在不同层次上的组织的结果,包括氨基酸序列、蛋白质结构、蛋白质组装和全层次微观结构。然而,相对较少的报告包含对序列全长的基序的分析,以及当纤维受到机械载荷时它们的二级结构的演变,这使得关联该材料的序列、微观结构和特性的任务变得困难。本研究鉴定了 Argiope bruennichi 的 7 种蜘蛛大安藻腺丝蛋白,确定了它们的全长氨基酸基序,模拟了蜘蛛大苯藻安藻腺丝 (MAS) 在自然环境中的反复拉伸,验证了其力学性能的稳定性,并通过拉曼光谱半定量分析建立了其蛋白质结构的演变。在不同应变下拉伸后,MAS 可以恢复以前的机械行为,并在纵向拉伸方面表现出优异的形状和机械记忆。研究还表明,MAS 通过精确调整蛋白质结构、二级结构转化和重建来保持其机械性能。本研究表明,蜘蛛大安瓿腺丝的反复拉伸特性可能是一种后处理调整方式,结合其分子组织,可能为序列-结构-性质的关系带来新的启发。
更新日期:2024-12-06
中文翻译:

使用基于 spidrion 分子生物信息学的拉曼光谱分析装载-卸载循环中的蜘蛛丝
蜘蛛丝的机械性能是其在不同层次上的组织的结果,包括氨基酸序列、蛋白质结构、蛋白质组装和全层次微观结构。然而,相对较少的报告包含对序列全长的基序的分析,以及当纤维受到机械载荷时它们的二级结构的演变,这使得关联该材料的序列、微观结构和特性的任务变得困难。本研究鉴定了 Argiope bruennichi 的 7 种蜘蛛大安藻腺丝蛋白,确定了它们的全长氨基酸基序,模拟了蜘蛛大苯藻安藻腺丝 (MAS) 在自然环境中的反复拉伸,验证了其力学性能的稳定性,并通过拉曼光谱半定量分析建立了其蛋白质结构的演变。在不同应变下拉伸后,MAS 可以恢复以前的机械行为,并在纵向拉伸方面表现出优异的形状和机械记忆。研究还表明,MAS 通过精确调整蛋白质结构、二级结构转化和重建来保持其机械性能。本研究表明,蜘蛛大安瓿腺丝的反复拉伸特性可能是一种后处理调整方式,结合其分子组织,可能为序列-结构-性质的关系带来新的启发。






























 京公网安备 11010802027423号
京公网安备 11010802027423号