Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Microcrystal Growth Pathways Investigated with Machine Learning Segmentation and Classification in Scanning Electron Microscopy
ACS Nano ( IF 15.8 ) Pub Date : 2024-11-19 , DOI: 10.1021/acsnano.4c08955 Rachel R. Chan, Jacob Pietryga, Kaitlin M. Landy, Kyle J. Gibson, Chad A. Mirkin
ACS Nano ( IF 15.8 ) Pub Date : 2024-11-19 , DOI: 10.1021/acsnano.4c08955 Rachel R. Chan, Jacob Pietryga, Kaitlin M. Landy, Kyle J. Gibson, Chad A. Mirkin
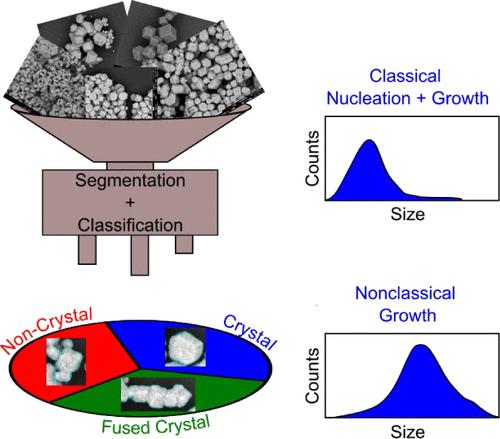
|
Advances in electron microscopy have revolutionized material characterization on the nano- and microscales, providing important insights into local ordering, structure, and size and quality distributions. While shape and size can be rigorously quantified through microscopy, it is often limited to local structure analysis and fails to describe bulk sample quality. Herein, a flexible machine learning (ML) tool is described that can segment and classify faceted crystals in scanning electron microscopy (SEM) micrographs to determine sample quality through the crystal size and product distribution. As a case study, this tool was applied to investigate crystal growth pathways (classical nucleation and growth compared to nonclassical growth) in DNA-mediated nanoparticle assembly through size and product (single crystal, fused crystal, or noncrystal) distribution of samples containing over 13000 colloidal crystal products. Strong DNA bond strengths (controlled by DNA sequence) lead to fast nucleation that exhausts the monomer concentration, resulting in smaller colloidal crystals. Alternatively, increased thermal energy and crystallization time lead to nonclassical crystallization pathways (coalescence) that result in larger colloidal crystals. This tool is useful since experimental conditions can now be deliberately identified to control colloidal crystal size and size distribution, important considerations for researchers interested in designing and synthesizing colloidal crystal metamaterials.
中文翻译:

在扫描电子显微镜中使用机器学习分割和分类研究微晶生长途径
电子显微镜的进步彻底改变了纳米和微米尺度上的材料表征,为局部排序、结构、尺寸和质量分布提供了重要的见解。虽然可以通过显微镜严格量化形状和大小,但它通常仅限于局部结构分析,无法描述散装样品的质量。本文描述了一种灵活的机器学习 (ML) 工具,该工具可以在扫描电子显微镜 (SEM) 显微照片中对多面晶体进行分割和分类,以通过晶体尺寸和产品分布确定样品质量。作为一个案例研究,该工具被用于研究 DNA 介导的纳米颗粒组装中的晶体生长途径(经典成核和生长与非经典生长相比),通过包含 13000 多种胶体晶体产物的样品的大小和产品(单晶、熔融晶体或非晶体)分布。强大的 DNA 键强度(由 DNA 序列控制)导致快速成核,耗尽单体浓度,从而产生更小的胶体晶体。或者,增加的热能和结晶时间会导致非经典结晶途径(聚结),从而导致更大的胶体晶体。该工具非常有用,因为现在可以有意识地确定实验条件来控制胶体晶体的尺寸和尺寸分布,这对于有兴趣设计和合成胶体晶体超材料的研究人员来说是重要的考虑因素。
更新日期:2024-11-20
中文翻译:

在扫描电子显微镜中使用机器学习分割和分类研究微晶生长途径
电子显微镜的进步彻底改变了纳米和微米尺度上的材料表征,为局部排序、结构、尺寸和质量分布提供了重要的见解。虽然可以通过显微镜严格量化形状和大小,但它通常仅限于局部结构分析,无法描述散装样品的质量。本文描述了一种灵活的机器学习 (ML) 工具,该工具可以在扫描电子显微镜 (SEM) 显微照片中对多面晶体进行分割和分类,以通过晶体尺寸和产品分布确定样品质量。作为一个案例研究,该工具被用于研究 DNA 介导的纳米颗粒组装中的晶体生长途径(经典成核和生长与非经典生长相比),通过包含 13000 多种胶体晶体产物的样品的大小和产品(单晶、熔融晶体或非晶体)分布。强大的 DNA 键强度(由 DNA 序列控制)导致快速成核,耗尽单体浓度,从而产生更小的胶体晶体。或者,增加的热能和结晶时间会导致非经典结晶途径(聚结),从而导致更大的胶体晶体。该工具非常有用,因为现在可以有意识地确定实验条件来控制胶体晶体的尺寸和尺寸分布,这对于有兴趣设计和合成胶体晶体超材料的研究人员来说是重要的考虑因素。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号