当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Highly Specific and Rapid Multiplex Identification of Candida Species Using Digital Microfluidics Integrated with a Semi-Nested Genoarray
Analytical Chemistry ( IF 6.7 ) Pub Date : 2024-11-16 , DOI: 10.1021/acs.analchem.4c04265 Zeyin Mao, Anni Deng, Xiangyu Jin, Tianqi Zhou, Shuailong Zhang, Meng Li, Wenqi Lv, Leyang Huang, Hao Zhong, Shihong Wang, Yixuan Shi, Lei Zhang, Qinping Liao, Rongxin Fu, Guoliang Huang
Analytical Chemistry ( IF 6.7 ) Pub Date : 2024-11-16 , DOI: 10.1021/acs.analchem.4c04265 Zeyin Mao, Anni Deng, Xiangyu Jin, Tianqi Zhou, Shuailong Zhang, Meng Li, Wenqi Lv, Leyang Huang, Hao Zhong, Shihong Wang, Yixuan Shi, Lei Zhang, Qinping Liao, Rongxin Fu, Guoliang Huang
|
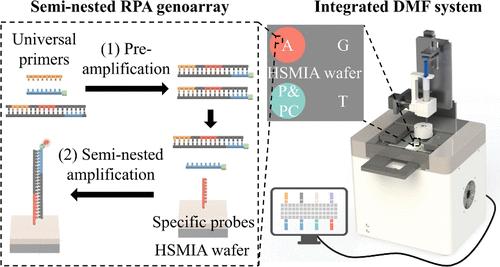
Candida species are the most common cause of fungal infections around the world, associated with superficial and even deep-seated infections. In clinical practice, there is great significance in identifying different Candida species because of their respective characteristics. However, current technologies have difficulty in onsite species identification due to long turnover time, high cost of reagents and instruments, or limited detection performance. We developed a semi-nested recombinase polymerase amplification (RPA) genoarray as well as an integrated system for highly specific identification of four Candida species with a simple design of primers, high detection sensitivity, fast turnover time, and good cost-effectiveness. The system constructed to perform the assay consists of a rapid sample processing module for nucleic acid release from fungal samples in 15 min and a digital microfluidic platform for precise and efficient detection reactions in 35 min. Therefore, our system could automatically identify specific Candida species, with a reagent consumption of only 2.5 μL of the RPA reaction mixture per target and no cross-reaction. Its detection sensitivity for four Candida species achieved 101–102 CFU/mL, which was 10-fold better than conventional RPA and even comparable to a common polymerase chain reaction. Evaluated by using cultured samples and 24 clinical samples, our system shows great applicability to onsite multiplex nucleic acid analysis.
中文翻译:

使用与半巢式基因阵列集成的数字微流控对念珠菌属进行高度特异性和快速的多重鉴定
念珠菌属是世界各地真菌感染的最常见原因,与浅表甚至深部感染有关。在临床实践中,由于不同的念珠菌属具有各自的特征,因此鉴定它们具有重要意义。然而,由于周转时间长、试剂和仪器成本高或检测性能有限,目前的技术难以进行现场物种鉴定。我们开发了一种半巢式重组酶聚合酶扩增 (RPA) 基因阵列以及一种用于高度特异性鉴定四种念珠菌属的集成系统,引物设计简单,检测灵敏度高,周转时间快,成本效益好。为执行测定而构建的系统包括一个快速样品处理模块,用于在 15 分钟内从真菌样品中释放核酸,以及一个数字微流控平台,用于在 35 分钟内进行精确和高效的检测反应。因此,我们的系统可以自动识别特定的念珠菌种类,每个靶标的试剂消耗量仅为 2.5 μL RPA 反应混合物,且无交叉反应。其对四种念珠菌属的检测灵敏度达到 101-10 2 CFU/mL,比传统 RPA 高 10 倍,甚至与普通聚合酶链反应相当。通过使用培养样本和 24 份临床样本进行评估,我们的系统显示出对现场多重核酸分析的极好适用性。
更新日期:2024-11-16
中文翻译:

使用与半巢式基因阵列集成的数字微流控对念珠菌属进行高度特异性和快速的多重鉴定
念珠菌属是世界各地真菌感染的最常见原因,与浅表甚至深部感染有关。在临床实践中,由于不同的念珠菌属具有各自的特征,因此鉴定它们具有重要意义。然而,由于周转时间长、试剂和仪器成本高或检测性能有限,目前的技术难以进行现场物种鉴定。我们开发了一种半巢式重组酶聚合酶扩增 (RPA) 基因阵列以及一种用于高度特异性鉴定四种念珠菌属的集成系统,引物设计简单,检测灵敏度高,周转时间快,成本效益好。为执行测定而构建的系统包括一个快速样品处理模块,用于在 15 分钟内从真菌样品中释放核酸,以及一个数字微流控平台,用于在 35 分钟内进行精确和高效的检测反应。因此,我们的系统可以自动识别特定的念珠菌种类,每个靶标的试剂消耗量仅为 2.5 μL RPA 反应混合物,且无交叉反应。其对四种念珠菌属的检测灵敏度达到 101-10 2 CFU/mL,比传统 RPA 高 10 倍,甚至与普通聚合酶链反应相当。通过使用培养样本和 24 份临床样本进行评估,我们的系统显示出对现场多重核酸分析的极好适用性。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号