Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
The single-molecule accessibility landscape of newly replicated mammalian chromatin
Cell ( IF 45.5 ) Pub Date : 2024-11-15 , DOI: 10.1016/j.cell.2024.10.039 Megan S. Ostrowski, Marty G. Yang, Colin P. McNally, Nour J. Abdulhay, Simai Wang, Keerthi Renduchintala, Iryna Irkliyenko, Alva Biran, Brandon T.L. Chew, Ayush D. Midha, Emily V. Wong, Jonathan Sandoval, Isha H. Jain, Anja Groth, Elphège P. Nora, Hani Goodarzi, Vijay Ramani
Cell ( IF 45.5 ) Pub Date : 2024-11-15 , DOI: 10.1016/j.cell.2024.10.039 Megan S. Ostrowski, Marty G. Yang, Colin P. McNally, Nour J. Abdulhay, Simai Wang, Keerthi Renduchintala, Iryna Irkliyenko, Alva Biran, Brandon T.L. Chew, Ayush D. Midha, Emily V. Wong, Jonathan Sandoval, Isha H. Jain, Anja Groth, Elphège P. Nora, Hani Goodarzi, Vijay Ramani
|
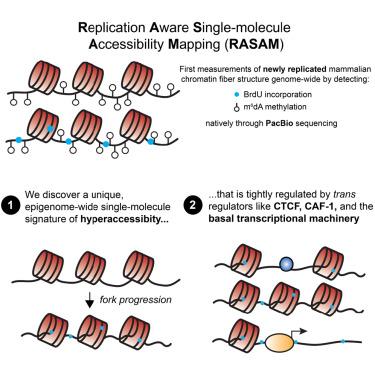
We present replication-aware single-molecule accessibility mapping (RASAM), a method to nondestructively measure replication status and protein-DNA interactions on chromatin genome-wide. Using RASAM, we uncover a genome-wide state of single-molecule “hyperaccessibility” post-replication that resolves over several hours. Combining RASAM with cellular models for rapid protein degradation, we demonstrate that histone chaperone CAF-1 reduces nascent chromatin accessibility by filling single-molecular “gaps” and generating closely spaced dinucleosomes on replicated DNA. At cis-regulatory elements, we observe unique modes by which nascent chromatin hyperaccessibility resolves: at CCCTC-binding factor (CTCF)-binding sites, CTCF and nucleosomes compete, reducing CTCF occupancy and motif accessibility post-replication; at active transcription start sites, high chromatin accessibility is maintained, implying rapid re-establishment of nucleosome-free regions. Our study introduces a new paradigm for studying replicated chromatin fiber organization. More broadly, we uncover a unique organization of newly replicated chromatin that must be reset by active processes, providing a substrate for epigenetic reprogramming.
中文翻译:

新复制的哺乳动物染色质的单分子可及性景观
我们提出了复制感知单分子可及性映射 (RASAM),这是一种无损测量全基因组染色质上复制状态和蛋白质-DNA 相互作用的方法。使用 RASAM,我们发现了复制后单分子“超可访问性”的全基因组状态,该状态在几个小时内解决。将 RASAM 与用于快速蛋白质降解的细胞模型相结合,我们证明组蛋白伴侣 CAF-1 通过填充单分子“间隙”并在复制的 DNA 上产生紧密间隔的二核小体来降低新生染色质的可及性。在顺式调节元件处,我们观察到新生染色质超可及性解决的独特模式:在 CCCTC 结合因子 (CTCF) 结合位点,CTCF 和核小体竞争,降低 CTCF 占有率和复制后基序可及性;在活跃的转录起始位点,保持了高染色质可及性,这意味着快速重建无核小体区域。我们的研究引入了一种研究复制的染色质纤维组织的新范式。更广泛地说,我们发现了新复制的染色质的独特组织,该组织必须通过活性过程重置,从而为表观遗传重编程提供底物。
更新日期:2024-11-15
中文翻译:

新复制的哺乳动物染色质的单分子可及性景观
我们提出了复制感知单分子可及性映射 (RASAM),这是一种无损测量全基因组染色质上复制状态和蛋白质-DNA 相互作用的方法。使用 RASAM,我们发现了复制后单分子“超可访问性”的全基因组状态,该状态在几个小时内解决。将 RASAM 与用于快速蛋白质降解的细胞模型相结合,我们证明组蛋白伴侣 CAF-1 通过填充单分子“间隙”并在复制的 DNA 上产生紧密间隔的二核小体来降低新生染色质的可及性。在顺式调节元件处,我们观察到新生染色质超可及性解决的独特模式:在 CCCTC 结合因子 (CTCF) 结合位点,CTCF 和核小体竞争,降低 CTCF 占有率和复制后基序可及性;在活跃的转录起始位点,保持了高染色质可及性,这意味着快速重建无核小体区域。我们的研究引入了一种研究复制的染色质纤维组织的新范式。更广泛地说,我们发现了新复制的染色质的独特组织,该组织必须通过活性过程重置,从而为表观遗传重编程提供底物。






























 京公网安备 11010802027423号
京公网安备 11010802027423号