Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Spatiotemporal modeling of molecular holograms
Cell ( IF 45.5 ) Pub Date : 2024-11-11 , DOI: 10.1016/j.cell.2024.10.011 Xiaojie Qiu, Daniel Y. Zhu, Yifan Lu, Jiajun Yao, Zehua Jing, Kyung Hoi Min, Mengnan Cheng, Hailin Pan, Lulu Zuo, Samuel King, Qi Fang, Huiwen Zheng, Mingyue Wang, Shuai Wang, Qingquan Zhang, Sichao Yu, Sha Liao, Chao Liu, Xinchao Wu, Yiwei Lai, Shijie Hao, Zhewei Zhang, Liang Wu, Yong Zhang, Mei Li, Zhencheng Tu, Jinpei Lin, Zhuoxuan Yang, Yuxiang Li, Ying Gu, David Ellison, Ao Chen, Longqi Liu, Jonathan S. Weissman, Jiayi Ma, Xun Xu, Shiping Liu, Yinqi Bai
Cell ( IF 45.5 ) Pub Date : 2024-11-11 , DOI: 10.1016/j.cell.2024.10.011 Xiaojie Qiu, Daniel Y. Zhu, Yifan Lu, Jiajun Yao, Zehua Jing, Kyung Hoi Min, Mengnan Cheng, Hailin Pan, Lulu Zuo, Samuel King, Qi Fang, Huiwen Zheng, Mingyue Wang, Shuai Wang, Qingquan Zhang, Sichao Yu, Sha Liao, Chao Liu, Xinchao Wu, Yiwei Lai, Shijie Hao, Zhewei Zhang, Liang Wu, Yong Zhang, Mei Li, Zhencheng Tu, Jinpei Lin, Zhuoxuan Yang, Yuxiang Li, Ying Gu, David Ellison, Ao Chen, Longqi Liu, Jonathan S. Weissman, Jiayi Ma, Xun Xu, Shiping Liu, Yinqi Bai
|
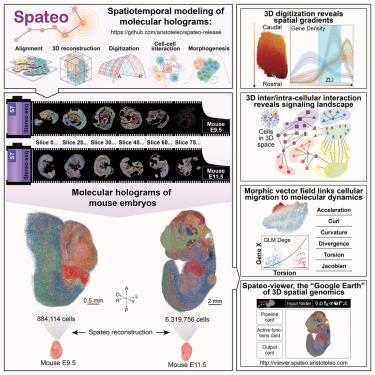
Quantifying spatiotemporal dynamics during embryogenesis is crucial for understanding congenital diseases. We developed Spateo (https://github.com/aristoteo/spateo-release ), a 3D spatiotemporal modeling framework, and applied it to a 3D mouse embryogenesis atlas at E9.5 and E11.5, capturing eight million cells. Spateo enables scalable, partial, non-rigid alignment, multi-slice refinement, and mesh correction to create molecular holograms of whole embryos. It introduces digitization methods to uncover multi-level biology from subcellular to whole organ, identifying expression gradients along orthogonal axes of emergent 3D structures, e.g., secondary organizers such as midbrain-hindbrain boundary (MHB). Spateo further jointly models intercellular and intracellular interaction to dissect signaling landscapes in 3D structures, including the zona limitans intrathalamica (ZLI). Lastly, Spateo introduces “morphometric vector fields” of cell migration and integrates spatial differential geometry to unveil molecular programs underlying asymmetrical murine heart organogenesis and others, bridging macroscopic changes with molecular dynamics. Thus, Spateo enables the study of organ ecology at a molecular level in 3D space over time.
中文翻译:

分子全息图的时空建模
量化胚胎发生过程中的时空动态对于理解先天性疾病至关重要。我们开发了 3D 时空建模框架 Spateo (https://github.com/aristoteo/spateo-release),并将其应用于 E9.5 和 E11.5 的 3D 小鼠胚胎发生图谱,捕获了 800 万个细胞。Spateo 支持可扩展、部分、非刚性对齐、多切片细化和网格校正,以创建整个胚胎的分子全息图。它引入了数字化方法,以揭示从亚细胞到整个器官的多层次生物学,识别沿新兴 3D 结构的正交轴的表达梯度,例如,中脑-后脑边界 (MHB) 等二级组织者。Spateo 进一步联合模拟细胞间和细胞内相互作用,以剖析 3D 结构中的信号传导景观,包括丘脑内限制带 (ZLI)。最后,Spateo 引入了细胞迁移的“形态测量向量场”,并整合了空间差分几何学,以揭示不对称小鼠心脏器官发生等的分子程序,将宏观变化与分子动力学联系起来。因此,Spateo 能够在 3D 空间中随时间推移在分子水平上研究器官生态学。
更新日期:2024-11-11
中文翻译:

分子全息图的时空建模
量化胚胎发生过程中的时空动态对于理解先天性疾病至关重要。我们开发了 3D 时空建模框架 Spateo (https://github.com/aristoteo/spateo-release),并将其应用于 E9.5 和 E11.5 的 3D 小鼠胚胎发生图谱,捕获了 800 万个细胞。Spateo 支持可扩展、部分、非刚性对齐、多切片细化和网格校正,以创建整个胚胎的分子全息图。它引入了数字化方法,以揭示从亚细胞到整个器官的多层次生物学,识别沿新兴 3D 结构的正交轴的表达梯度,例如,中脑-后脑边界 (MHB) 等二级组织者。Spateo 进一步联合模拟细胞间和细胞内相互作用,以剖析 3D 结构中的信号传导景观,包括丘脑内限制带 (ZLI)。最后,Spateo 引入了细胞迁移的“形态测量向量场”,并整合了空间差分几何学,以揭示不对称小鼠心脏器官发生等的分子程序,将宏观变化与分子动力学联系起来。因此,Spateo 能够在 3D 空间中随时间推移在分子水平上研究器官生态学。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号