当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
AMARO: All Heavy-Atom Transferable Neural Network Potentials of Protein Thermodynamics
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2024-11-08 , DOI: 10.1021/acs.jctc.4c01239 Antonio Mirarchi, Raúl P. Peláez, Guillem Simeon, Gianni De Fabritiis
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2024-11-08 , DOI: 10.1021/acs.jctc.4c01239 Antonio Mirarchi, Raúl P. Peláez, Guillem Simeon, Gianni De Fabritiis
|
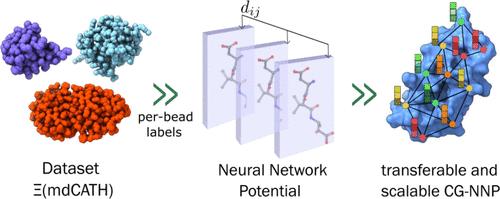
All-atom molecular simulations offer detailed insights into macromolecular phenomena, but their substantial computational cost hinders the exploration of complex biological processes. We introduce Advanced Machine-learning Atomic Representation Omni-force-field (AMARO), a new neural network potential (NNP) that combines an O(3)-equivariant message-passing neural network architecture, TensorNet, with a coarse-graining map that excludes hydrogen atoms. AMARO demonstrates the feasibility of training coarser NNP, without prior energy terms, to run stable protein dynamics with scalability and generalization capabilities.
中文翻译:

AMARO:蛋白质热力学的所有重原子可传递神经网络势
全原子分子模拟提供了对大分子现象的详细见解,但其大量的计算成本阻碍了对复杂生物过程的探索。我们介绍了高级机器学习原子表示全力场 (AMARO),这是一种新的神经网络势 (NNP),它结合了 O(3) 等变消息传递神经网络架构 TensorNet 和排除氢原子的粗粒度映射。AMARO 展示了在没有先验能量项的情况下训练较粗的 NNP 以运行具有可扩展性和泛化能力的稳定蛋白质动力学的可行性。
更新日期:2024-11-08
中文翻译:

AMARO:蛋白质热力学的所有重原子可传递神经网络势
全原子分子模拟提供了对大分子现象的详细见解,但其大量的计算成本阻碍了对复杂生物过程的探索。我们介绍了高级机器学习原子表示全力场 (AMARO),这是一种新的神经网络势 (NNP),它结合了 O(3) 等变消息传递神经网络架构 TensorNet 和排除氢原子的粗粒度映射。AMARO 展示了在没有先验能量项的情况下训练较粗的 NNP 以运行具有可扩展性和泛化能力的稳定蛋白质动力学的可行性。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号