当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Pred-AHCP: Robust Feature Selection-Enabled Sequence-Specific Prediction of Anti-Hepatitis C Peptides via Machine Learning
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-11-06 , DOI: 10.1021/acs.jcim.4c00900 Akash Saraswat, Utsav Sharma, Aryan Gandotra, Lakshit Wasan, Sainithin Artham, Arijit Maitra, Bipin Singh
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-11-06 , DOI: 10.1021/acs.jcim.4c00900 Akash Saraswat, Utsav Sharma, Aryan Gandotra, Lakshit Wasan, Sainithin Artham, Arijit Maitra, Bipin Singh
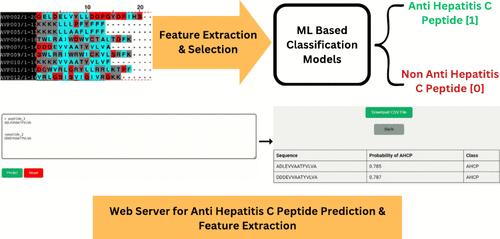
|
Every year, an estimated 1.5 million people worldwide contract Hepatitis C, a significant contributor to liver problems. Although many studies have explored machine learning’s potential to predict antiviral peptides, very few have addressed the problem of predicting peptides against specific viruses such as Hepatitis C. In this study, we demonstrate the application and fine-tuning of machine learning (ML) algorithms to predict peptides that are effective against Hepatitis C virus (HCV). We developed a fine-tuned and explainable ML model that harnesses the amino acid sequence of a peptide to predict its anti-hepatitis C potential. Specifically, features were computed based on sequence and physicochemical properties. The feature selection was performed using a combined strategy of mutual information and variance inflation factor. This facilitated the removal of redundant and multicollinear features, enhancing the model’s generalizability in predicting anti-hepatitis C peptides (AHCPs). The model using the random forest algorithm produced the best performance with an accuracy of about 92%. The feature analysis highlights that the distributions of hydrophobicity, polarizability, coil-forming residues, frequency of glycine residues and the existence of dipeptide motifs VL, LV, and CC emerged as the key predictors for identifying AHCPs targeting different components of HCV. The developed model can be accessed through the Pred-AHCP web server, provided at http://tinyurl.com/web-Pred-AHCP. This resource facilitates the prediction and re-engineering of AHCPs for designing peptide-based therapeutics while also proposing an exploration of similar strategies for designing peptide inhibitors effective against other viruses. The developed ML model can also be used for validating peptide sequences generated using generative artificial intelligence methods for further optimization.
中文翻译:

Pred-AHCP:通过机器学习对抗丙型肝炎肽进行稳健特征选择的序列特异性预测
每年,全世界估计有 150 万人感染丙型肝炎,丙型肝炎是导致肝脏问题的重要因素。尽管许多研究已经探索了机器学习预测抗病毒肽的潜力,但很少有研究解决了预测针对特定病毒(如丙型肝炎)的肽的问题。在这项研究中,我们展示了机器学习 (ML) 算法的应用和微调,以预测对丙型肝炎病毒 (HCV) 有效的肽。我们开发了一个微调且可解释的 ML 模型,该模型利用肽的氨基酸序列来预测其抗丙型肝炎潜力。具体来说,特征是根据序列和物理化学性质计算的。特征选择是使用互信息和方差膨胀因子的组合策略进行的。这有助于去除冗余和多重共线特征,增强了模型在预测抗丙型肝炎肽 (AHCP) 方面的普遍性。使用随机森林算法的模型产生了最佳性能,准确率约为 92%。特征分析强调,疏水性、极化性、线圈形成残基、甘氨酸残基的频率以及二肽基序 VL 、 LV 和 CC 的存在成为鉴定靶向 HCV 不同组分的 AHCP 的关键预测因子。开发的模型可以通过 http://tinyurl.com/web-Pred-AHCP 提供的 Pred-AHCP Web 服务器访问。该资源有助于预测和重新设计 AHCP 以设计基于肽的疗法,同时还提出了设计对其他病毒有效的肽抑制剂的类似策略的探索。 开发的 ML 模型还可用于验证使用生成式人工智能方法生成的肽序列,以进一步优化。
更新日期:2024-11-07
中文翻译:

Pred-AHCP:通过机器学习对抗丙型肝炎肽进行稳健特征选择的序列特异性预测
每年,全世界估计有 150 万人感染丙型肝炎,丙型肝炎是导致肝脏问题的重要因素。尽管许多研究已经探索了机器学习预测抗病毒肽的潜力,但很少有研究解决了预测针对特定病毒(如丙型肝炎)的肽的问题。在这项研究中,我们展示了机器学习 (ML) 算法的应用和微调,以预测对丙型肝炎病毒 (HCV) 有效的肽。我们开发了一个微调且可解释的 ML 模型,该模型利用肽的氨基酸序列来预测其抗丙型肝炎潜力。具体来说,特征是根据序列和物理化学性质计算的。特征选择是使用互信息和方差膨胀因子的组合策略进行的。这有助于去除冗余和多重共线特征,增强了模型在预测抗丙型肝炎肽 (AHCP) 方面的普遍性。使用随机森林算法的模型产生了最佳性能,准确率约为 92%。特征分析强调,疏水性、极化性、线圈形成残基、甘氨酸残基的频率以及二肽基序 VL 、 LV 和 CC 的存在成为鉴定靶向 HCV 不同组分的 AHCP 的关键预测因子。开发的模型可以通过 http://tinyurl.com/web-Pred-AHCP 提供的 Pred-AHCP Web 服务器访问。该资源有助于预测和重新设计 AHCP 以设计基于肽的疗法,同时还提出了设计对其他病毒有效的肽抑制剂的类似策略的探索。 开发的 ML 模型还可用于验证使用生成式人工智能方法生成的肽序列,以进一步优化。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号