当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Accelerated Simulations Reveal Physicochemical Factors Governing Stability and Composition of RNA Clusters
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2024-11-06 , DOI: 10.1021/acs.jctc.4c00803 Dilimulati Aierken, Jerelle A. Joseph
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2024-11-06 , DOI: 10.1021/acs.jctc.4c00803 Dilimulati Aierken, Jerelle A. Joseph
|
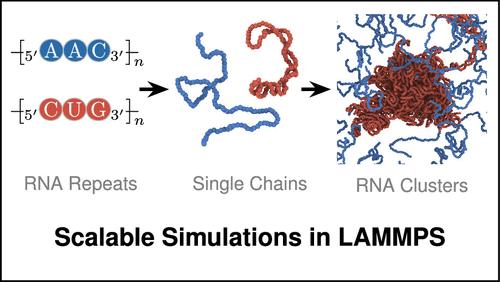
Under certain conditions, RNA repeat sequences phase separate, yielding protein-free biomolecular condensates. Importantly, RNA repeat sequences have also been implicated in neurological disorders, such as Huntington’s disease. Thus, mapping repeat sequences to their phase behavior, functions, and dysfunctions is an active area of research. However, despite several advances, it remains challenging to characterize the RNA phase behavior at a submolecular resolution. Here, we have implemented a residue-resolution coarse-grained model in LAMMPS─that incorporates both the RNA sequence and structure─to study the clustering propensities of protein-free RNA systems. Importantly, we achieve a multifold speedup in the simulation time compared to previous work. Leveraging this efficiency, we study the clustering propensity of all 20 nonredundant trinucleotide repeat sequences. Our results align with findings from experiments, emphasizing that canonical base-pairing and G–U wobble pairs play dominant roles in regulating cluster formation of RNA repeat sequences. Strikingly, we find strong entropic contributions to the stability and composition of RNA clusters, which is demonstrated for single-component RNA systems as well as binary mixtures of trinucleotide repeats. Additionally, we investigate the clustering behaviors of trinucleotide (odd) repeats and their quadranucleotide (even) counterparts. We observe that odd repeats exhibit stronger clustering tendencies, attributed to the presence of consecutive base pairs in their sequences that are disrupted in even repeat sequences. Altogether, our work extends the set of computational tools for probing RNA cluster formation at submolecular resolution and uncovers physicochemical principles that govern the stability and composition of the resulting clusters.
中文翻译:

加速模拟揭示了控制 RNA 簇稳定性和组成的理化因素
在某些条件下,RNA 重复序列相分离,产生无蛋白质的生物分子缩合物。重要的是,RNA 重复序列也与神经系统疾病有关,例如亨廷顿病。因此,将重复序列映射到它们的相位行为、功能和功能障碍是一个活跃的研究领域。然而,尽管取得了一些进展,但在亚分子分辨率下表征 RNA 相行为仍然具有挑战性。在这里,我们在 LAMMPS 中实现了一个残基分辨率粗粒度模型——它结合了 RNA 序列和结构——来研究无蛋白质 RNA 系统的聚类倾向。重要的是,与以前的工作相比,我们的仿真时间加快了数倍。利用这种效率,我们研究了所有 20 个非冗余三核苷酸重复序列的聚类倾向。我们的结果与实验结果一致,强调经典碱基配对和 G-U 摆动对在调节 RNA 重复序列的簇形成中起主导作用。引人注目的是,我们发现熵对 RNA 簇的稳定性和组成有很强的贡献,这在单组分 RNA 系统以及三核苷酸重复序列的二元混合物中得到了证明。此外,我们研究了三核苷酸 (奇数) 重复序列及其四核苷酸 (偶数) 对应物的聚类行为。我们观察到奇数重复表现出更强的聚类趋势,这归因于其序列中存在连续的碱基对,这些碱基对在偶数重复序列中被破坏。 总而言之,我们的工作扩展了以亚分子分辨率探测 RNA 簇形成的计算工具集,并揭示了控制所得簇稳定性和组成的物理化学原理。
更新日期:2024-11-07
中文翻译:

加速模拟揭示了控制 RNA 簇稳定性和组成的理化因素
在某些条件下,RNA 重复序列相分离,产生无蛋白质的生物分子缩合物。重要的是,RNA 重复序列也与神经系统疾病有关,例如亨廷顿病。因此,将重复序列映射到它们的相位行为、功能和功能障碍是一个活跃的研究领域。然而,尽管取得了一些进展,但在亚分子分辨率下表征 RNA 相行为仍然具有挑战性。在这里,我们在 LAMMPS 中实现了一个残基分辨率粗粒度模型——它结合了 RNA 序列和结构——来研究无蛋白质 RNA 系统的聚类倾向。重要的是,与以前的工作相比,我们的仿真时间加快了数倍。利用这种效率,我们研究了所有 20 个非冗余三核苷酸重复序列的聚类倾向。我们的结果与实验结果一致,强调经典碱基配对和 G-U 摆动对在调节 RNA 重复序列的簇形成中起主导作用。引人注目的是,我们发现熵对 RNA 簇的稳定性和组成有很强的贡献,这在单组分 RNA 系统以及三核苷酸重复序列的二元混合物中得到了证明。此外,我们研究了三核苷酸 (奇数) 重复序列及其四核苷酸 (偶数) 对应物的聚类行为。我们观察到奇数重复表现出更强的聚类趋势,这归因于其序列中存在连续的碱基对,这些碱基对在偶数重复序列中被破坏。 总而言之,我们的工作扩展了以亚分子分辨率探测 RNA 簇形成的计算工具集,并揭示了控制所得簇稳定性和组成的物理化学原理。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号