当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
StaPep: An Open-Source Toolkit for Structure Prediction, Feature Extraction, and Rational Design of Hydrocarbon-Stapled Peptides
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-11-06 , DOI: 10.1021/acs.jcim.4c01718 Zhe Wang, Jianping Wu, Mengjun Zheng, Chenchen Geng, Borui Zhen, Wei Zhang, Hui Wu, Zhengyang Xu, Gang Xu, Si Chen, Xiang Li
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-11-06 , DOI: 10.1021/acs.jcim.4c01718 Zhe Wang, Jianping Wu, Mengjun Zheng, Chenchen Geng, Borui Zhen, Wei Zhang, Hui Wu, Zhengyang Xu, Gang Xu, Si Chen, Xiang Li
|
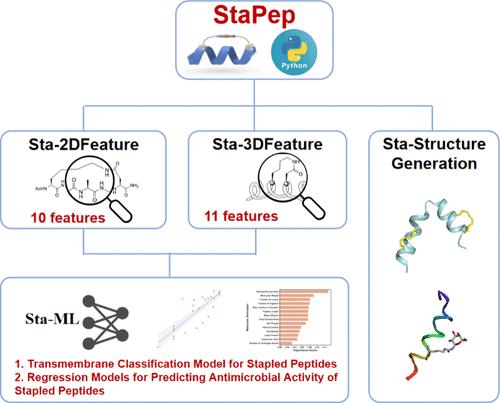
All-hydrocarbon stapled peptides, with their covalent side-chain constraints, provide enhanced proteolytic stability and membrane permeability, making them superior to linear peptides. However, tools for extracting structural and physicochemical descriptors to predict the properties of hydrocarbon-stapled peptides are lacking. To address this, we present StaPep, a Python-based toolkit for generating 3D structures and calculating 21 features for hydrocarbon-stapled peptides. StaPep supports peptides containing two non-standard amino acids (norleucine and 2-aminoisobutyric acid) and six non-natural anchoring residues (S3, S5, S8, R3, R5, and R8), with customization options for other non-standard amino acids. We showcase StaPep’s utility through three case studies. The first generates 3D structures of these peptides with a mean RMSD of 1.62 ± 0.86, offering essential structural insights for drug design and biological activity prediction. The second develops machine learning models based on calculated molecular features to differentiate between membrane-permeable and non-permeable stapled peptides, achieving an AUC of 0.93. The third constructs regression models to predict the antimicrobial activity of stapled peptides against Escherichia coli, with a Pearson correlation of 0.84. StaPep’s pipeline spans data retrieval, structure generation, feature calculation, and machine learning modeling for hydrocarbon-stapled peptides. The source codes and data set are freely available on Github: https://github.com/dahuilangda/stapep_package.
中文翻译:

StaPep:用于碳氢化合物装订肽结构预测、特征提取和合理设计的开源工具包
全烃类吻合肽具有共价侧链约束,可提供增强的蛋白水解稳定性和膜通透性,使其优于线性肽。然而,缺乏提取结构和物理化学描述符以预测碳氢化合物订书肽性质的工具。为了解决这个问题,我们提出了 StaPep,这是一个基于 Python 的工具包,用于生成 3D 结构和计算碳氢化合物吻合肽的 21 个特征。StaPep 支持含有两种非标准氨基酸(正亮氨酸和 2-氨基异丁酸)和六种非天然锚定残基(S3、S5、S8、R3、R5 和 R8)的肽,并提供其他非标准氨基酸的定制选项。我们通过三个案例研究展示了 StaPep 的效用。第一个生成这些肽的 3D 结构,平均 RMSD 为 1.62 ± 0.86,为药物设计和生物活性预测提供重要的结构见解。第二个项目基于计算出的分子特征开发机器学习模型,以区分膜渗透性和非渗透性吻合肽,AUC 为 0.93。第三个构建回归模型来预测装订肽对大肠杆菌的抗菌活性,Pearson 相关性为 0.84。StataPep 的流程涵盖碳氢化合物吻合肽的数据检索、结构生成、特征计算和机器学习建模。源代码和数据集可在 Github 上免费获得:https://github.com/dahuilangda/stapep_package。
更新日期:2024-11-07
中文翻译:

StaPep:用于碳氢化合物装订肽结构预测、特征提取和合理设计的开源工具包
全烃类吻合肽具有共价侧链约束,可提供增强的蛋白水解稳定性和膜通透性,使其优于线性肽。然而,缺乏提取结构和物理化学描述符以预测碳氢化合物订书肽性质的工具。为了解决这个问题,我们提出了 StaPep,这是一个基于 Python 的工具包,用于生成 3D 结构和计算碳氢化合物吻合肽的 21 个特征。StaPep 支持含有两种非标准氨基酸(正亮氨酸和 2-氨基异丁酸)和六种非天然锚定残基(S3、S5、S8、R3、R5 和 R8)的肽,并提供其他非标准氨基酸的定制选项。我们通过三个案例研究展示了 StaPep 的效用。第一个生成这些肽的 3D 结构,平均 RMSD 为 1.62 ± 0.86,为药物设计和生物活性预测提供重要的结构见解。第二个项目基于计算出的分子特征开发机器学习模型,以区分膜渗透性和非渗透性吻合肽,AUC 为 0.93。第三个构建回归模型来预测装订肽对大肠杆菌的抗菌活性,Pearson 相关性为 0.84。StataPep 的流程涵盖碳氢化合物吻合肽的数据检索、结构生成、特征计算和机器学习建模。源代码和数据集可在 Github 上免费获得:https://github.com/dahuilangda/stapep_package。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号