当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
CACHE Challenge #1: Targeting the WDR Domain of LRRK2, A Parkinson’s Disease Associated Protein
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-11-05 , DOI: 10.1021/acs.jcim.4c01267 Fengling Li, Suzanne Ackloo, Cheryl H. Arrowsmith, Fuqiang Ban, Christopher J. Barden, Hartmut Beck, Jan Beránek, Francois Berenger, Albina Bolotokova, Guillaume Bret, Marko Breznik, Emanuele Carosati, Irene Chau, Yu Chen, Artem Cherkasov, Dennis Della Corte, Katrin Denzinger, Aiping Dong, Sorin Draga, Ian Dunn, Kristina Edfeldt, Aled Edwards, Merveille Eguida, Paul Eisenhuth, Lukas Friedrich, Alexander Fuerll, Spencer S Gardiner, Francesco Gentile, Pegah Ghiabi, Elisa Gibson, Marta Glavatskikh, Christoph Gorgulla, Judith Guenther, Anders Gunnarsson, Filipp Gusev, Evgeny Gutkin, Levon Halabelian, Rachel J. Harding, Alexander Hillisch, Laurent Hoffer, Anders Hogner, Scott Houliston, John J Irwin, Olexandr Isayev, Aleksandra Ivanova, Celien Jacquemard, Austin J Jarrett, Jan H. Jensen, Dmitri Kireev, Julian Kleber, S. Benjamin Koby, David Koes, Ashutosh Kumar, Maria G. Kurnikova, Alina Kutlushina, Uta Lessel, Fabian Liessmann, Sijie Liu, Wei Lu, Jens Meiler, Akhila Mettu, Guzel Minibaeva, Rocco Moretti, Connor J Morris, Chamali Narangoda, Theresa Noonan, Leon Obendorf, Szymon Pach, Amit Pandit, Sumera Perveen, Gennady Poda, Pavel Polishchuk, Kristina Puls, Vera Pütter, Didier Rognan, Dylan Roskams-Edris, Christina Schindler, François Sindt, Vojtěch Spiwok, Casper Steinmann, Rick L. Stevens, Valerij Talagayev, Damon Tingey, Oanh Vu, W. Patrick Walters, Xiaowen Wang, Zhenyu Wang, Gerhard Wolber, Clemens Alexander Wolf, Lars Wortmann, Hong Zeng, Carlos A. Zepeda, Kam Y. J. Zhang, Jixian Zhang, Shuangjia Zheng, Matthieu Schapira
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-11-05 , DOI: 10.1021/acs.jcim.4c01267 Fengling Li, Suzanne Ackloo, Cheryl H. Arrowsmith, Fuqiang Ban, Christopher J. Barden, Hartmut Beck, Jan Beránek, Francois Berenger, Albina Bolotokova, Guillaume Bret, Marko Breznik, Emanuele Carosati, Irene Chau, Yu Chen, Artem Cherkasov, Dennis Della Corte, Katrin Denzinger, Aiping Dong, Sorin Draga, Ian Dunn, Kristina Edfeldt, Aled Edwards, Merveille Eguida, Paul Eisenhuth, Lukas Friedrich, Alexander Fuerll, Spencer S Gardiner, Francesco Gentile, Pegah Ghiabi, Elisa Gibson, Marta Glavatskikh, Christoph Gorgulla, Judith Guenther, Anders Gunnarsson, Filipp Gusev, Evgeny Gutkin, Levon Halabelian, Rachel J. Harding, Alexander Hillisch, Laurent Hoffer, Anders Hogner, Scott Houliston, John J Irwin, Olexandr Isayev, Aleksandra Ivanova, Celien Jacquemard, Austin J Jarrett, Jan H. Jensen, Dmitri Kireev, Julian Kleber, S. Benjamin Koby, David Koes, Ashutosh Kumar, Maria G. Kurnikova, Alina Kutlushina, Uta Lessel, Fabian Liessmann, Sijie Liu, Wei Lu, Jens Meiler, Akhila Mettu, Guzel Minibaeva, Rocco Moretti, Connor J Morris, Chamali Narangoda, Theresa Noonan, Leon Obendorf, Szymon Pach, Amit Pandit, Sumera Perveen, Gennady Poda, Pavel Polishchuk, Kristina Puls, Vera Pütter, Didier Rognan, Dylan Roskams-Edris, Christina Schindler, François Sindt, Vojtěch Spiwok, Casper Steinmann, Rick L. Stevens, Valerij Talagayev, Damon Tingey, Oanh Vu, W. Patrick Walters, Xiaowen Wang, Zhenyu Wang, Gerhard Wolber, Clemens Alexander Wolf, Lars Wortmann, Hong Zeng, Carlos A. Zepeda, Kam Y. J. Zhang, Jixian Zhang, Shuangjia Zheng, Matthieu Schapira
|
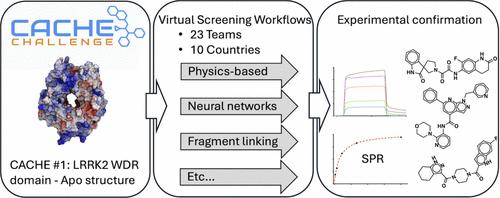
The CACHE challenges are a series of prospective benchmarking exercises to evaluate progress in the field of computational hit-finding. Here we report the results of the inaugural CACHE challenge in which 23 computational teams each selected up to 100 commercially available compounds that they predicted would bind to the WDR domain of the Parkinson’s disease target LRRK2, a domain with no known ligand and only an apo structure in the PDB. The lack of known binding data and presumably low druggability of the target is a challenge to computational hit finding methods. Of the 1955 molecules predicted by participants in Round 1 of the challenge, 73 were found to bind to LRRK2 in an SPR assay with a KD lower than 150 μM. These 73 molecules were advanced to the Round 2 hit expansion phase, where computational teams each selected up to 50 analogs. Binding was observed in two orthogonal assays for seven chemically diverse series, with affinities ranging from 18 to 140 μM. The seven successful computational workflows varied in their screening strategies and techniques. Three used molecular dynamics to produce a conformational ensemble of the targeted site, three included a fragment docking step, three implemented a generative design strategy and five used one or more deep learning steps. CACHE #1 reflects a highly exploratory phase in computational drug design where participants adopted strikingly diverging screening strategies. Machine learning-accelerated methods achieved similar results to brute force (e.g., exhaustive) docking. First-in-class, experimentally confirmed compounds were rare and weakly potent, indicating that recent advances are not sufficient to effectively address challenging targets.
中文翻译:

CACHE 挑战 #1:靶向帕金森病相关蛋白 LRRK2 的 WDR 结构域
CACHE 挑战是一系列前瞻性基准测试练习,用于评估计算命中查找领域的进展。在这里,我们报告了首届 CACHE 挑战赛的结果,在该挑战赛中,23 个计算团队各自选择了多达 100 种市售化合物,他们预测这些化合物将与帕金森病靶标 LRRK2 的 WDR 结构域结合,该结构域在 PDB 中没有已知的配体,只有一个 apo 结构。缺乏已知的结合数据和靶标的低成药性是对计算命中查找方法的挑战。在第 1 轮挑战赛参与者预测的 1955 个分子中,发现 73 个分子在 SPR 测定中与 LRRK2 结合,KD 低于 150 μM。这 73 个分子进入了第 2 轮命中扩展阶段,每个计算团队最多选择了 50 个类似物。在 7 个化学不同系列的 2 次正交测定中观察到结合,亲和力范围为 18 至 140 μM。7 个成功的计算工作流程在筛选策略和技术方面有所不同。3 项研究使用分子动力学生成目标位点的构象集合,3 项研究包括片段对接步骤,3 项研究实施了生成式设计策略,5 项研究使用一个或多个深度学习步骤。CACHE #1 反映了计算药物设计中的一个高度探索性阶段,参与者采用了截然不同的筛选策略。机器学习加速方法取得了与蛮力(例如,穷举)对接相似的结果。经实验证实的同类首创化合物稀有且效力较弱,这表明最近的进展不足以有效解决具有挑战性的靶标。
更新日期:2024-11-06
中文翻译:

CACHE 挑战 #1:靶向帕金森病相关蛋白 LRRK2 的 WDR 结构域
CACHE 挑战是一系列前瞻性基准测试练习,用于评估计算命中查找领域的进展。在这里,我们报告了首届 CACHE 挑战赛的结果,在该挑战赛中,23 个计算团队各自选择了多达 100 种市售化合物,他们预测这些化合物将与帕金森病靶标 LRRK2 的 WDR 结构域结合,该结构域在 PDB 中没有已知的配体,只有一个 apo 结构。缺乏已知的结合数据和靶标的低成药性是对计算命中查找方法的挑战。在第 1 轮挑战赛参与者预测的 1955 个分子中,发现 73 个分子在 SPR 测定中与 LRRK2 结合,KD 低于 150 μM。这 73 个分子进入了第 2 轮命中扩展阶段,每个计算团队最多选择了 50 个类似物。在 7 个化学不同系列的 2 次正交测定中观察到结合,亲和力范围为 18 至 140 μM。7 个成功的计算工作流程在筛选策略和技术方面有所不同。3 项研究使用分子动力学生成目标位点的构象集合,3 项研究包括片段对接步骤,3 项研究实施了生成式设计策略,5 项研究使用一个或多个深度学习步骤。CACHE #1 反映了计算药物设计中的一个高度探索性阶段,参与者采用了截然不同的筛选策略。机器学习加速方法取得了与蛮力(例如,穷举)对接相似的结果。经实验证实的同类首创化合物稀有且效力较弱,这表明最近的进展不足以有效解决具有挑战性的靶标。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号