当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Exploring Free Energy Landscapes for Protein Partitioning into Membrane Domains in All-Atom and Coarse-Grained Simulations
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2024-11-01 , DOI: 10.1021/acs.jctc.4c00881 Seulki Kwon, Ayan Majumder, John E. Straub
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2024-11-01 , DOI: 10.1021/acs.jctc.4c00881 Seulki Kwon, Ayan Majumder, John E. Straub
|
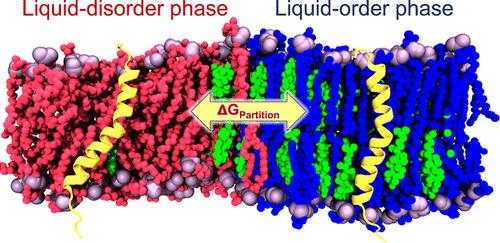
It is known that membrane environment can impact the structure and function of integral membrane proteins. As such, elucidation of the thermodynamic driving forces governing protein partitioning between membrane domains of varying lipid composition is a fundamental topic in membrane biophysics. Molecular dynamics simulations provide valuable tools for quantitatively characterizing the free energy landscapes governing protein partitioning at the molecular level. In this study, we propose an efficient simulation methodology for the calculation of free energies for the partitioning of transmembrane proteins between liquid-disorder (Ld) and liquid-ordered (Lo) domains in all-atom (AA) phase-separated lipid bilayers. The computed potential of mean force defining the equilibrium partition coefficients is compared for AA and coarse-grained systems. Energy decomposition is used to identify differences in the underlying thermodynamics. Our findings highlight the importance of employing AA models to accurately estimate relevant free energy changes during protein translation between membrane domains.
中文翻译:

在全原子和粗粒度模拟中探索蛋白质分配到膜结构域的自由能景观
众所周知,膜环境会影响整合膜蛋白的结构和功能。因此,阐明控制不同脂质组成的膜结构域之间蛋白质分配的热力学驱动力是膜生物物理学的一个基本主题。分子动力学模拟为定量表征分子水平控制蛋白质分配的自由能景观提供了有价值的工具。在这项研究中,我们提出了一种有效的模拟方法,用于计算全原子 (AA) 相分离脂质双层中液体无序 (Ld) 和液体有序 (Lo) 结构域之间跨膜蛋白分配的自由能。比较了定义平衡分配系数的平均力的计算势能,用于 AA 和粗粒度系统。能量分解用于识别潜在热力学的差异。我们的研究结果强调了使用 AA 模型准确估计膜结构域之间蛋白质翻译过程中相关自由能变化的重要性。
更新日期:2024-11-01
中文翻译:

在全原子和粗粒度模拟中探索蛋白质分配到膜结构域的自由能景观
众所周知,膜环境会影响整合膜蛋白的结构和功能。因此,阐明控制不同脂质组成的膜结构域之间蛋白质分配的热力学驱动力是膜生物物理学的一个基本主题。分子动力学模拟为定量表征分子水平控制蛋白质分配的自由能景观提供了有价值的工具。在这项研究中,我们提出了一种有效的模拟方法,用于计算全原子 (AA) 相分离脂质双层中液体无序 (Ld) 和液体有序 (Lo) 结构域之间跨膜蛋白分配的自由能。比较了定义平衡分配系数的平均力的计算势能,用于 AA 和粗粒度系统。能量分解用于识别潜在热力学的差异。我们的研究结果强调了使用 AA 模型准确估计膜结构域之间蛋白质翻译过程中相关自由能变化的重要性。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号