当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
AlphaFold Meets De Novo Drug Design: Leveraging Structural Protein Information in Multitarget Molecular Generative Models
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-30 , DOI: 10.1021/acs.jcim.4c00309 Andrius Bernatavicius, Martin Šícho, Antonius P. A. Janssen, Alan Kai Hassen, Mike Preuss, Gerard J. P. van Westen
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-30 , DOI: 10.1021/acs.jcim.4c00309 Andrius Bernatavicius, Martin Šícho, Antonius P. A. Janssen, Alan Kai Hassen, Mike Preuss, Gerard J. P. van Westen
|
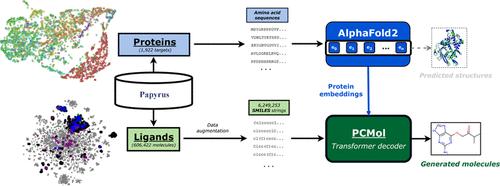
Recent advancements in deep learning and generative models have significantly expanded the applications of virtual screening for drug-like compounds. Here, we introduce a multitarget transformer model, PCMol, that leverages the latent protein embeddings derived from AlphaFold2 as a means of conditioning a de novo generative model on different targets. Incorporating rich protein representations allows the model to capture their structural relationships, enabling the chemical space interpolation of active compounds and target-side generalization to new proteins based on embedding similarities. In this work, we benchmark against other existing target-conditioned transformer models to illustrate the validity of using AlphaFold protein representations over raw amino acid sequences. We show that low-dimensional projections of these protein embeddings cluster appropriately based on target families and that model performance declines when these representations are intentionally corrupted. We also show that the PCMol model generates diverse, potentially active molecules for a wide array of proteins, including those with sparse ligand bioactivity data. The generated compounds display higher similarity known active ligands of held-out targets and have comparable molecular docking scores while maintaining novelty. Additionally, we demonstrate the important role of data augmentation in bolstering the performance of generative models in low-data regimes. Software package and AlphaFold protein embeddings are freely available at https://github.com/CDDLeiden/PCMol.
中文翻译:

AlphaFold 与从头药物设计相遇:在多靶点分子生成模型中利用结构蛋白信息
深度学习和生成模型的最新进展显著扩展了药物样化合物虚拟筛选的应用。在这里,我们介绍了一个多靶点转换器模型 PCMol,它利用源自 AlphaFold2 的潜在蛋白质嵌入作为在不同靶标上调节从头生成模型的手段。结合丰富的蛋白质表示使模型能够捕获它们的结构关系,从而能够对活性化合物进行化学空间插值,并根据嵌入相似性对新蛋白质进行靶标侧泛化。在这项工作中,我们与其他现有的靶标条件 transformer 模型进行基准测试,以说明在原始氨基酸序列上使用 AlphaFold 蛋白表示的有效性。我们表明,这些蛋白质嵌入的低维投影根据目标家族适当地聚集,并且当这些表示被故意破坏时,模型性能会下降。我们还表明,PCMol 模型为多种蛋白质生成多种潜在活性分子,包括那些具有稀疏配体生物活性数据的蛋白质。生成的化合物与保留靶标的已知活性配体具有更高的相似性,并且在保持新颖性的同时具有可比的分子对接分数。此外,我们还展示了数据增强在提高生成模型在低数据状态下的性能方面的重要作用。软件包和 AlphaFold 蛋白包埋可在 https://github.com/CDDLeiden/PCMol 免费获得。
更新日期:2024-10-30
中文翻译:

AlphaFold 与从头药物设计相遇:在多靶点分子生成模型中利用结构蛋白信息
深度学习和生成模型的最新进展显著扩展了药物样化合物虚拟筛选的应用。在这里,我们介绍了一个多靶点转换器模型 PCMol,它利用源自 AlphaFold2 的潜在蛋白质嵌入作为在不同靶标上调节从头生成模型的手段。结合丰富的蛋白质表示使模型能够捕获它们的结构关系,从而能够对活性化合物进行化学空间插值,并根据嵌入相似性对新蛋白质进行靶标侧泛化。在这项工作中,我们与其他现有的靶标条件 transformer 模型进行基准测试,以说明在原始氨基酸序列上使用 AlphaFold 蛋白表示的有效性。我们表明,这些蛋白质嵌入的低维投影根据目标家族适当地聚集,并且当这些表示被故意破坏时,模型性能会下降。我们还表明,PCMol 模型为多种蛋白质生成多种潜在活性分子,包括那些具有稀疏配体生物活性数据的蛋白质。生成的化合物与保留靶标的已知活性配体具有更高的相似性,并且在保持新颖性的同时具有可比的分子对接分数。此外,我们还展示了数据增强在提高生成模型在低数据状态下的性能方面的重要作用。软件包和 AlphaFold 蛋白包埋可在 https://github.com/CDDLeiden/PCMol 免费获得。

































 京公网安备 11010802027423号
京公网安备 11010802027423号