当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integrated Knowledge Graph and Drug Molecular Graph Fusion via Adversarial Networks for Drug–Drug Interaction Prediction
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-30 , DOI: 10.1021/acs.jcim.4c01647 Yu Li, Zhu-Hong You, Yang Yuan, Cheng-Gang Mi, Yu-An Huang, Hai-Cheng Yi, Lin-Xuan Hou
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-30 , DOI: 10.1021/acs.jcim.4c01647 Yu Li, Zhu-Hong You, Yang Yuan, Cheng-Gang Mi, Yu-An Huang, Hai-Cheng Yi, Lin-Xuan Hou
|
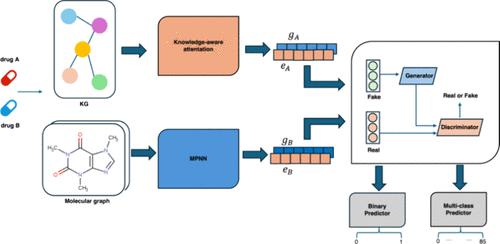
The Co-administration of multiple drugs can enhance the efficacy of disease treatment by reducing drug resistance and side effects. However, it also raises the risk of adverse drug interactions, presenting a challenging problem in healthcare. Various approaches have been developed to predict drug–drug interactions (DDIs) by leveraging both knowledge graphs and drug attribute information. While these methods have shown promise, they often fail to effectively capture correlations between biomedical information in the knowledge graph and drug properties. This work introduces a novel end-to-end DDI predictor framework based on generative adversarial networks. This framework utilizes a message-passing neural network to capture molecular structure information while employing the knowledge-aware graph attention network to capture the representation of drugs in the knowledge graph through considering the importance of different multihop neighbor nodes and relationships. The dual generative adversarial networks employ two generators and two discriminators in knowledge graph embedding and molecular topology embedding for adversarial training to capture the interrelations and complementary knowledge between molecular structure information and semantic information from the knowledge graph. comprehensive experiments have demonstrated that the proposed method outperforms state-of-the-art algorithms in binary classification, with improvements of 1.0% in accuracy, 0.45% in area under the receiver operating characteristic curve (AUC), 0.24% in area under the precision-recall curve (AUPR), and 0.98% in F1 score. Furthermore, for multiclass classification tasks, improvements were observed across various evaluation metrics, including 0.9% in accuracy, 0.25% in macro precision, 0.2% in macro recall, and 0.28% in macro F1. Additionally, ablation studies were conducted to showcase the effectiveness and robustness of our method in DDI prediction tasks.
中文翻译:

通过对抗网络集成知识图谱和药物分子图谱融合,用于药物相互作用预测
多种药物的联合给药可以通过减少耐药性和副作用来提高疾病治疗的疗效。然而,它也增加了药物不良反应的风险,在医疗保健领域提出了一个具有挑战性的问题。已经开发了各种方法,通过利用知识图谱和药物属性信息来预测药物相互作用 (DDI)。虽然这些方法已显示出前景,但它们通常无法有效地捕捉知识图谱中生物医学信息与药物特性之间的相关性。这项工作介绍了一种基于生成对抗网络的新型端到端 DDI 预测器框架。该框架利用消息传递神经网络来捕获分子结构信息,同时通过考虑不同多跳邻居节点和关系的重要性,采用知识感知图注意力网络来捕获药物在知识图谱中的表示。双生成对抗网络在知识图谱嵌入和分子拓扑嵌入中采用两个生成器和两个判别器进行对抗训练,以捕获知识图谱中分子结构信息和语义信息之间的相互关系和互补知识。综合实验表明,所提出的方法在二元分类方面优于最先进的算法,准确率提高了 1.0%,受试者工作特征曲线下面积 (AUC) 提高了 0.45%,精度-召回曲线下面积 (AUPR) 提高了 0.24%,F1 评分提高了 0.98%。此外,对于多类分类任务,在各种评估指标上都观察到改进,包括准确率 0.9%,宏观精度 0.25%,0。宏观召回率为 2%,宏观 F1 为 0.28%。此外,还进行了消融研究,以展示我们的方法在 DDI 预测任务中的有效性和稳健性。
更新日期:2024-10-30
中文翻译:

通过对抗网络集成知识图谱和药物分子图谱融合,用于药物相互作用预测
多种药物的联合给药可以通过减少耐药性和副作用来提高疾病治疗的疗效。然而,它也增加了药物不良反应的风险,在医疗保健领域提出了一个具有挑战性的问题。已经开发了各种方法,通过利用知识图谱和药物属性信息来预测药物相互作用 (DDI)。虽然这些方法已显示出前景,但它们通常无法有效地捕捉知识图谱中生物医学信息与药物特性之间的相关性。这项工作介绍了一种基于生成对抗网络的新型端到端 DDI 预测器框架。该框架利用消息传递神经网络来捕获分子结构信息,同时通过考虑不同多跳邻居节点和关系的重要性,采用知识感知图注意力网络来捕获药物在知识图谱中的表示。双生成对抗网络在知识图谱嵌入和分子拓扑嵌入中采用两个生成器和两个判别器进行对抗训练,以捕获知识图谱中分子结构信息和语义信息之间的相互关系和互补知识。综合实验表明,所提出的方法在二元分类方面优于最先进的算法,准确率提高了 1.0%,受试者工作特征曲线下面积 (AUC) 提高了 0.45%,精度-召回曲线下面积 (AUPR) 提高了 0.24%,F1 评分提高了 0.98%。此外,对于多类分类任务,在各种评估指标上都观察到改进,包括准确率 0.9%,宏观精度 0.25%,0。宏观召回率为 2%,宏观 F1 为 0.28%。此外,还进行了消融研究,以展示我们的方法在 DDI 预测任务中的有效性和稳健性。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号