当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Unravelling the Complexity of Amyloid Peptide Core Interfaces
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-29 , DOI: 10.1021/acs.jcim.4c01479 Máté Sulyok-Eiler, Veronika Harmat, András Perczel
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-29 , DOI: 10.1021/acs.jcim.4c01479 Máté Sulyok-Eiler, Veronika Harmat, András Perczel
|
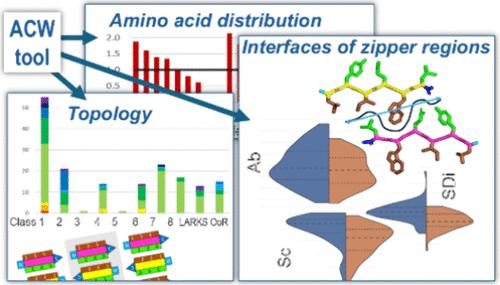
Amyloids, large intermolecular sandwiched β-sheet structures, underlie several protein misfolding diseases but have also been shown to have functional roles and can be a basis for designing smart and responsive nanomaterials. Short segments of proteins, called aggregation-prone regions (APRs), have been identified that nucleate amyloid formation. Here we present the database of 173 APR crystal structures currently available in the PDB, and a tool, ACW, for analyzing their topologies and the 267 inter-β-sheet interfaces of zipper regions assigned in these structures. We defined a new descriptor of zipper interfaces, the surface detail index (SDi), which quantifies the intertwining between the side chains of both β-sheets of the zipper, an important factor for the molecular recognition and self-assembly of these mesostructures. This allowed a comparative analysis of the zipper interfaces and identification of 6 clusters with different intertwining, steric fit, and size characteristics using three complementary descriptors, SDi, shape complementarity, and buried surface area. 60% of the APR structures are formed by parallel β-sheets, of which 52% belong to the topological class 1. This could be explained by the better fit and a deeper entanglement of the zipper regions of the parallel structures than of the antiparallel structures, as the analysis showed that both their shape complementarity (0.79 vs 0.70) and SDi (1.53 vs 1.32) were higher. The higher abundance of certain residues (Asn and Gln in parallel and Leu and Ala in antiparallel β-sheets) can be explained by their ability to form different ladder-like secondary interaction patterns within β-sheets. Analogous to the hierarchy of protein structure, we interpreted the primary, secondary, tertiary, and quaternary structure levels of APRs revealing different characteristics of the zipper regions for both parallel and antiparallel β-sheet structures, which may provide clues to the structural conditions of amyloid core formation and the rational design of amyloid polymorphs.
中文翻译:

揭示淀粉样肽核心界面的复杂性
淀粉样蛋白是大型分子间夹β片结构,是几种蛋白质错误折叠疾病的基础,但也已被证明具有功能作用,可以成为设计智能和响应式纳米材料的基础。已经确定蛋白质的短片段,称为聚集易聚集区域 (APR),可以成核淀粉样蛋白形成。在这里,我们展示了 PDB 中当前可用的 173 种 APR 晶体结构的数据库,以及用于分析它们的拓扑结构以及这些结构中分配的拉链区域的 267 个β片界面的工具 ACW。我们定义了拉链界面的新描述符,即表面细节指数 (SDi),它量化了拉链两张β片侧链之间的交织,这是这些介观结构的分子识别和自组装的重要因素。这允许对拉链界面进行比较分析,并使用三个互补描述符 SDi、形状互补性和埋藏表面积识别具有不同交织、空间拟合和尺寸特征的 6 个簇。60% 的 APR 结构由平行β片形成,其中 52% 属于拓扑类 1。这可以通过平行结构的拉链区域比反平行结构更好的拟合和更深的纠缠来解释,因为分析表明它们的形状互补性(0.79 对 0.70)和 SDi(1.53 对 1.32)都更高。某些残基(Asn 和 Gln 平行,Leu 和 Ala 在反平行β片)的较高丰度可以通过它们在 β 片内形成不同的梯状次级相互作用模式的能力来解释。 类似于蛋白质结构的层次结构,我们解释了 APRs 的一级、二级、三级和四级结构水平,揭示了平行和反平行 β 片结构的拉链区域的不同特征,这可能为淀粉样蛋白核心形成的结构条件和淀粉样蛋白多晶型物的合理设计提供线索。
更新日期:2024-10-30
中文翻译:

揭示淀粉样肽核心界面的复杂性
淀粉样蛋白是大型分子间夹β片结构,是几种蛋白质错误折叠疾病的基础,但也已被证明具有功能作用,可以成为设计智能和响应式纳米材料的基础。已经确定蛋白质的短片段,称为聚集易聚集区域 (APR),可以成核淀粉样蛋白形成。在这里,我们展示了 PDB 中当前可用的 173 种 APR 晶体结构的数据库,以及用于分析它们的拓扑结构以及这些结构中分配的拉链区域的 267 个β片界面的工具 ACW。我们定义了拉链界面的新描述符,即表面细节指数 (SDi),它量化了拉链两张β片侧链之间的交织,这是这些介观结构的分子识别和自组装的重要因素。这允许对拉链界面进行比较分析,并使用三个互补描述符 SDi、形状互补性和埋藏表面积识别具有不同交织、空间拟合和尺寸特征的 6 个簇。60% 的 APR 结构由平行β片形成,其中 52% 属于拓扑类 1。这可以通过平行结构的拉链区域比反平行结构更好的拟合和更深的纠缠来解释,因为分析表明它们的形状互补性(0.79 对 0.70)和 SDi(1.53 对 1.32)都更高。某些残基(Asn 和 Gln 平行,Leu 和 Ala 在反平行β片)的较高丰度可以通过它们在 β 片内形成不同的梯状次级相互作用模式的能力来解释。 类似于蛋白质结构的层次结构,我们解释了 APRs 的一级、二级、三级和四级结构水平,揭示了平行和反平行 β 片结构的拉链区域的不同特征,这可能为淀粉样蛋白核心形成的结构条件和淀粉样蛋白多晶型物的合理设计提供线索。

































 京公网安备 11010802027423号
京公网安备 11010802027423号