当前位置:
X-MOL 学术
›
Anal. Chim. Acta
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Rapid and accurate bacteria identification through deep-learning-based two-dimensional Raman spectroscopy
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2024-10-29 , DOI: 10.1016/j.aca.2024.343376 Yichen Liu, Yisheng Gao, Rui Niu, Zunyue Zhang, Guo-Wei Lu, Haofeng Hu, Tiegen Liu, Zhenzhou Cheng
Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2024-10-29 , DOI: 10.1016/j.aca.2024.343376 Yichen Liu, Yisheng Gao, Rui Niu, Zunyue Zhang, Guo-Wei Lu, Haofeng Hu, Tiegen Liu, Zhenzhou Cheng
|
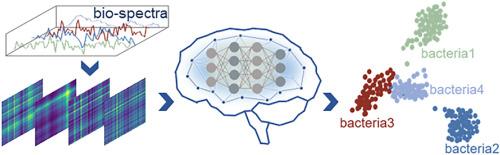
Surface-enhanced Raman spectroscopy (SERS) offers a distinctive vibrational fingerprint of the molecules and has led to widespread applications in medical diagnosis, biochemistry, and virology. With the rapid development of artificial intelligence (AI) technology, AI-enabled Raman spectroscopic techniques, as a promising avenue for biosensing applications, have significantly boosted bacteria identification. By converting spectra into images, the dataset is enriched with more detailed information, allowing AI to identify bacterial isolates with enhanced precision. However, previous studies usually suffer from a trade-off between high-resolution spectrograms for high-accuracy identification and short training time for data processing. Here, we present an efficient bacteria identification strategy that combines deep learning models with a spectrogram encoding algorithm based on wavelet packet transform and Gramian angular field techniques. In contrast to the direct analysis of raw Raman spectra, our approach utilizes wavelet packet transform techniques to compress the spectra by a factor of 1/15, while concurrently maintaining state-of-the-art accuracy by amplifying the subtle differences via Gramian angular field techniques. The results demonstrate that our approach can achieve a 99.64 % and a 90.55 % identification accuracy for two types of bacterial isolates and thirty types of bacterial isolates, respectively, while a 90 % reduction in training time compared to the conventional methods. To verify the model's stability, Gaussian noises were superimposed on the testing dataset, showing a specific generalization ability and superior performance. This algorithm has the potential for integration into on-site testing protocols and is readily updatable with new bacterial isolates. This study provides profound insights and contributes to the current understanding of spectroscopy, paving the way for accurate and rapid bacteria identification in diverse applications of environment monitoring, food safety, microbiology, and public health.
中文翻译:

通过基于深度学习的二维拉曼光谱快速准确地鉴定细菌
表面增强拉曼光谱 (SERS) 提供了分子的独特振动指纹,并在医学诊断、生物化学和病毒学中得到了广泛的应用。随着人工智能 (AI) 技术的快速发展,人工智能拉曼光谱技术作为生物传感应用的前景广阔的途径,极大地促进了细菌鉴定。通过将光谱转换为图像,数据集中可以丰富更详细的信息,使 AI 能够以更高的精度识别细菌分离株。然而,以前的研究通常在用于高精度识别的高分辨率频谱图和用于数据处理的短训练时间之间进行权衡。在这里,我们提出了一种高效的细菌识别策略,该策略将深度学习模型与基于小波包变换和格拉米亚角场技术的频谱图编码算法相结合。与直接分析原始拉曼光谱相比,我们的方法利用小波包变换技术将光谱压缩 1/15 倍,同时通过格拉米亚角场技术放大细微差异,同时保持最先进的精度。结果表明,我们的方法可以分别实现 99.64% 和 90.55% 的两种细菌分离物和 30 种细菌分离物的识别准确率,同时与传统方法相比,训练时间减少了 90%。为了验证模型的稳定性,将高斯噪声叠加在测试数据集上,显示出特定的泛化能力和卓越的性能。 该算法有可能集成到现场检测方案中,并且很容易使用新的细菌分离物进行更新。这项研究提供了深刻的见解,并有助于当前对光谱学的理解,为在环境监测、食品安全、微生物学和公共卫生的各种应用中准确、快速地鉴定细菌铺平了道路。
更新日期:2024-10-29
中文翻译:

通过基于深度学习的二维拉曼光谱快速准确地鉴定细菌
表面增强拉曼光谱 (SERS) 提供了分子的独特振动指纹,并在医学诊断、生物化学和病毒学中得到了广泛的应用。随着人工智能 (AI) 技术的快速发展,人工智能拉曼光谱技术作为生物传感应用的前景广阔的途径,极大地促进了细菌鉴定。通过将光谱转换为图像,数据集中可以丰富更详细的信息,使 AI 能够以更高的精度识别细菌分离株。然而,以前的研究通常在用于高精度识别的高分辨率频谱图和用于数据处理的短训练时间之间进行权衡。在这里,我们提出了一种高效的细菌识别策略,该策略将深度学习模型与基于小波包变换和格拉米亚角场技术的频谱图编码算法相结合。与直接分析原始拉曼光谱相比,我们的方法利用小波包变换技术将光谱压缩 1/15 倍,同时通过格拉米亚角场技术放大细微差异,同时保持最先进的精度。结果表明,我们的方法可以分别实现 99.64% 和 90.55% 的两种细菌分离物和 30 种细菌分离物的识别准确率,同时与传统方法相比,训练时间减少了 90%。为了验证模型的稳定性,将高斯噪声叠加在测试数据集上,显示出特定的泛化能力和卓越的性能。 该算法有可能集成到现场检测方案中,并且很容易使用新的细菌分离物进行更新。这项研究提供了深刻的见解,并有助于当前对光谱学的理解,为在环境监测、食品安全、微生物学和公共卫生的各种应用中准确、快速地鉴定细菌铺平了道路。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号