当前位置:
X-MOL 学术
›
Cell Host Microbe
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Comprehensive analysis of Mycobacterium tuberculosis genomes reveals genetic variations in bacterial virulence
Cell Host & Microbe ( IF 20.6 ) Pub Date : 2024-10-28 , DOI: 10.1016/j.chom.2024.10.004 Wittawin Worakitchanon, Hideki Yanai, Pundharika Piboonsiri, Reiko Miyahara, Supalert Nedsuwan, Worarat Imsanguan, Boonchai Chaiyasirinroje, Waritta Sawaengdee, Sukanya Wattanapokayakit, Nuanjan Wichukchinda, Yosuke Omae, Prasit Palittapongarnpim, Katsushi Tokunaga, Surakameth Mahasirimongkol, Akihiro Fujimoto
Cell Host & Microbe ( IF 20.6 ) Pub Date : 2024-10-28 , DOI: 10.1016/j.chom.2024.10.004 Wittawin Worakitchanon, Hideki Yanai, Pundharika Piboonsiri, Reiko Miyahara, Supalert Nedsuwan, Worarat Imsanguan, Boonchai Chaiyasirinroje, Waritta Sawaengdee, Sukanya Wattanapokayakit, Nuanjan Wichukchinda, Yosuke Omae, Prasit Palittapongarnpim, Katsushi Tokunaga, Surakameth Mahasirimongkol, Akihiro Fujimoto
|
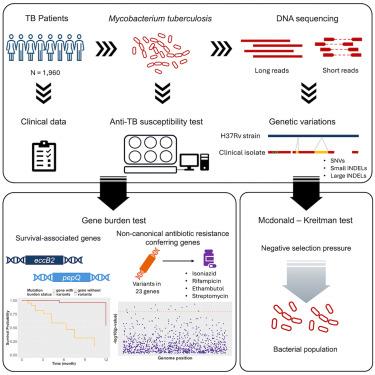
Tuberculosis, a disease caused by Mycobacterium tuberculosis (Mtb), is a significant health problem worldwide. Here, we developed a method to detect large insertions and deletions (indels), which have been generally understudied. Leveraging this framework, we performed a comprehensive analysis of single nucleotide variants and small and large indels across 1,960 Mtb clinical isolates. Comparing the distribution of variants demonstrated that gene disruptive variants are underrepresented in genes essential for bacterial survival. An evolutionary analysis revealed that Mtb genomes are enriched in partially deleterious mutations. Genome-wide association studies identified small and large deletions in eccB2 significantly associated with patient prognosis. Additionally, we unveil significant associations with antibiotic resistance in 23 non-canonical genes. Among these, large indels are primarily found in genetic regions of Rv1216c, Rv1217c, fadD11, and ctpD. This study provides a comprehensive catalog of genetic variations and highlights their potential impact for the future treatment and risk prediction of tuberculosis.
中文翻译:

结核分枝杆菌基因组的综合分析揭示了细菌毒力的遗传变异
结核病是一种由结核分枝杆菌 (Mtb) 引起的疾病,是世界范围内的一个重大健康问题。在这里,我们开发了一种检测大插入和缺失 (indel) 的方法,这些插入和缺失通常研究不足。利用这个框架,我们对 1,960 Mtb 临床分离株中的单核苷酸变异和小插入缺失和大插入缺失进行了全面分析。比较变异的分布表明,基因破坏性变异在细菌生存所必需的基因中代表性不足。进化分析显示,Mtb 基因组富含部分有害突变。全基因组关联研究确定了 eccB2 中的小缺失和大缺失与患者预后显著相关。此外,我们揭示了 23 个非经典基因与抗生素耐药性的显着关联。其中,大插入缺失主要存在于 Rv1216c 、 Rv1217c 、 fadD11 和 ctpD 的遗传区域。本研究提供了遗传变异的全面目录,并强调了它们对结核病未来治疗和风险预测的潜在影响。
更新日期:2024-10-28
中文翻译:

结核分枝杆菌基因组的综合分析揭示了细菌毒力的遗传变异
结核病是一种由结核分枝杆菌 (Mtb) 引起的疾病,是世界范围内的一个重大健康问题。在这里,我们开发了一种检测大插入和缺失 (indel) 的方法,这些插入和缺失通常研究不足。利用这个框架,我们对 1,960 Mtb 临床分离株中的单核苷酸变异和小插入缺失和大插入缺失进行了全面分析。比较变异的分布表明,基因破坏性变异在细菌生存所必需的基因中代表性不足。进化分析显示,Mtb 基因组富含部分有害突变。全基因组关联研究确定了 eccB2 中的小缺失和大缺失与患者预后显著相关。此外,我们揭示了 23 个非经典基因与抗生素耐药性的显着关联。其中,大插入缺失主要存在于 Rv1216c 、 Rv1217c 、 fadD11 和 ctpD 的遗传区域。本研究提供了遗传变异的全面目录,并强调了它们对结核病未来治疗和风险预测的潜在影响。






























 京公网安备 11010802027423号
京公网安备 11010802027423号