当前位置:
X-MOL 学术
›
Cell Chem. Bio.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Dynamic sampling of a surveillance state enables DNA proofreading by Cas9
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2024-10-28 , DOI: 10.1016/j.chembiol.2024.10.001 Viviane S. De Paula, Abhinav Dubey, Haribabu Arthanari, Nikolaos G. Sgourakis
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2024-10-28 , DOI: 10.1016/j.chembiol.2024.10.001 Viviane S. De Paula, Abhinav Dubey, Haribabu Arthanari, Nikolaos G. Sgourakis
|
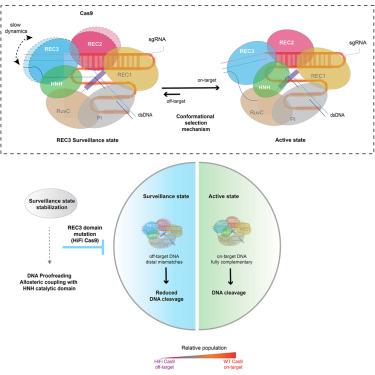
CRISPR-Cas9 has revolutionized genome engineering applications by programming its single-guide RNA, where high specificity is required. However, the precise molecular mechanism underscoring discrimination between on/off-target DNA sequences, relative to the guide RNA template, remains elusive. Here, using methyl-based NMR to study multiple holoenzymes assembled in vitro, we elucidate a discrete protein conformational state which enables recognition of DNA mismatches at the protospacer adjacent motif (PAM)-distal end. Our results delineate an allosteric pathway connecting a dynamic conformational switch at the REC3 domain, with the sampling of a catalytically competent state by the HNH domain. Our NMR data show that HiFi Cas9 (R691A) increases the fidelity of DNA recognition by stabilizing this "surveillance state" for mismatched substrates, shifting the Cas9 conformational equilibrium away from the active state. These results establish a paradigm of substrate recognition through an allosteric protein-based switch, providing unique insights into the molecular mechanism which governs Cas9 selectivity.
中文翻译:

监测状态的动态采样使 Cas9 能够进行 DNA 校对
CRISPR-Cas9 通过对其需要高特异性的单向导 RNA 进行编程,彻底改变了基因组工程应用。然而,相对于向导 RNA 模板,强调脱靶 DNA 序列之间区分的确切分子机制仍然难以捉摸。在这里,使用基于甲基的 NMR 研究体外 组装的多种全酶,我们阐明了一种离散的蛋白质构象状态,该状态能够识别前间隔区相邻基序 (PAM) 远端的 DNA 错配。我们的结果描绘了一条变构途径,该途径连接了 REC3 结构域的动态构象开关,以及 HNH 结构域对催化感受态的采样。我们的 NMR 数据显示,HiFi Cas9 (R691A) 通过稳定错配底物的这种“监视状态”,使 Cas9 构象平衡从活性状态转移出来,从而提高了 DNA 识别的保真度。这些结果通过基于变构蛋白的开关建立了底物识别的范式,为控制 Cas9 选择性的分子机制提供了独特的见解。
更新日期:2024-10-28
中文翻译:

监测状态的动态采样使 Cas9 能够进行 DNA 校对
CRISPR-Cas9 通过对其需要高特异性的单向导 RNA 进行编程,彻底改变了基因组工程应用。然而,相对于向导 RNA 模板,强调脱靶 DNA 序列之间区分的确切分子机制仍然难以捉摸。在这里,使用基于甲基的 NMR 研究体外 组装的多种全酶,我们阐明了一种离散的蛋白质构象状态,该状态能够识别前间隔区相邻基序 (PAM) 远端的 DNA 错配。我们的结果描绘了一条变构途径,该途径连接了 REC3 结构域的动态构象开关,以及 HNH 结构域对催化感受态的采样。我们的 NMR 数据显示,HiFi Cas9 (R691A) 通过稳定错配底物的这种“监视状态”,使 Cas9 构象平衡从活性状态转移出来,从而提高了 DNA 识别的保真度。这些结果通过基于变构蛋白的开关建立了底物识别的范式,为控制 Cas9 选择性的分子机制提供了独特的见解。































 京公网安备 11010802027423号
京公网安备 11010802027423号