Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
ANTIPASTI: Interpretable prediction of antibody binding affinity exploiting normal modes and deep learning
Structure ( IF 4.4 ) Pub Date : 2024-10-25 , DOI: 10.1016/j.str.2024.10.001 Kevin Michalewicz, Mauricio Barahona, Barbara Bravi
Structure ( IF 4.4 ) Pub Date : 2024-10-25 , DOI: 10.1016/j.str.2024.10.001 Kevin Michalewicz, Mauricio Barahona, Barbara Bravi
|
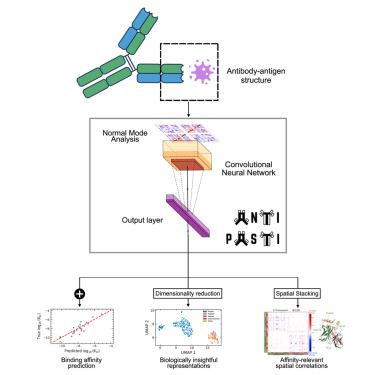
The high binding affinity of antibodies toward their cognate targets is key to eliciting effective immune responses, as well as to the use of antibodies as research and therapeutic tools. Here, we propose ANTIPASTI, a convolutional neural network model that achieves state-of-the-art performance in the prediction of antibody binding affinity using as input a representation of antibody-antigen structures in terms of normal mode correlation maps derived from elastic network models. This representation captures not only structural features but energetic patterns of local and global residue fluctuations. The learnt representations are interpretable: they reveal similarities of binding patterns among antibodies targeting the same antigen type, and can be used to quantify the importance of antibody regions contributing to binding affinity. Our results show the importance of the antigen imprint in the normal mode landscape, and the dominance of cooperative effects and long-range correlations between antibody regions to determine binding affinity.
中文翻译:

ANTIPASTI:利用正常模式和深度学习对抗体结合亲和力的可解释预测
抗体对其同源靶标的高结合亲和力是引发有效免疫反应的关键,也是使用抗体作为研究和治疗工具的关键。在这里,我们提出了 ANTIPASTI,这是一种卷积神经网络模型,它使用从弹性网络模型得出的正态模式相关图作为抗体-抗原结构的表示作为输入,在预测抗体结合亲和力方面实现了最先进的性能。这种表示不仅捕捉了结构特征,还捕捉了局部和全局残留物波动的能量模式。学习到的表示是可解释的:它们揭示了靶向相同抗原类型的抗体之间结合模式的相似性,并可用于量化有助于结合亲和力的抗体区域的重要性。我们的结果表明抗原印记在正常模式景观中的重要性,以及抗体区域之间协同效应和长程相关性在确定结合亲和力方面的优势。
更新日期:2024-10-25
中文翻译:

ANTIPASTI:利用正常模式和深度学习对抗体结合亲和力的可解释预测
抗体对其同源靶标的高结合亲和力是引发有效免疫反应的关键,也是使用抗体作为研究和治疗工具的关键。在这里,我们提出了 ANTIPASTI,这是一种卷积神经网络模型,它使用从弹性网络模型得出的正态模式相关图作为抗体-抗原结构的表示作为输入,在预测抗体结合亲和力方面实现了最先进的性能。这种表示不仅捕捉了结构特征,还捕捉了局部和全局残留物波动的能量模式。学习到的表示是可解释的:它们揭示了靶向相同抗原类型的抗体之间结合模式的相似性,并可用于量化有助于结合亲和力的抗体区域的重要性。我们的结果表明抗原印记在正常模式景观中的重要性,以及抗体区域之间协同效应和长程相关性在确定结合亲和力方面的优势。






























 京公网安备 11010802027423号
京公网安备 11010802027423号