当前位置:
X-MOL 学术
›
Chem. Rev.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Selection of Nucleotide-Encoded Mass Libraries of Macrocyclic Peptides for Inaccessible Drug Targets
Chemical Reviews ( IF 51.4 ) Pub Date : 2024-10-25 , DOI: 10.1021/acs.chemrev.4c00422 Kilian Colas, Daniel Bindl, Hiroaki Suga
Chemical Reviews ( IF 51.4 ) Pub Date : 2024-10-25 , DOI: 10.1021/acs.chemrev.4c00422 Kilian Colas, Daniel Bindl, Hiroaki Suga
|
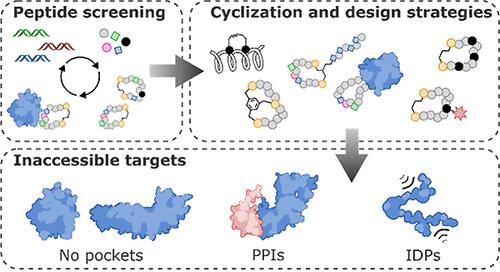
Technological advances and breakthrough developments in the pharmaceutical field are knocking at the door of the “undruggable” fortress with increasing insistence. Notably, the 21st century has seen the emergence of macrocyclic compounds, among which cyclic peptides are of particular interest. This new class of potential drug candidates occupies the vast chemical space between classic small-molecule drugs and larger protein-based therapeutics, such as antibodies. As research advances toward clinical targets that have long been considered inaccessible, macrocyclic peptides are well-suited to tackle these challenges in a post-rule of 5 pharmaceutical landscape. Facilitating their discovery is an arsenal of high-throughput screening methods that exploit massive randomized libraries of genetically encoded compounds. These techniques benefit from the incorporation of non-natural moieties, such as non- proteinogenic amino acids or stabilizing hydrocarbon staples. Exploiting these features for the strategic architectural design of macrocyclic peptides has the potential to tackle challenging targets such as protein–protein interactions, which have long resisted research efforts. This Review summarizes the basic principles and recent developments of the main high-throughput techniques for the discovery of macrocyclic peptides and focuses on their specific deployment for targeting undruggable space. A particular focus is placed on the development of new design guidelines and principles for the cyclization and structural stabilization of cyclic peptides and the resulting success stories achieved against well-known inaccessible drug targets.
中文翻译:

为难以接近的药物靶标选择核苷酸编码的大环肽质量库
制药领域的技术进步和突破性发展正在以越来越坚定的坚持敲响“不可成药”堡垒的大门。值得注意的是,21 世纪出现了大环化合物,其中环肽特别令人感兴趣。这类新型的潜在候选药物占据了经典小分子药物和大型蛋白质疗法(如抗体)之间的巨大化学空间。随着研究朝着长期以来被认为无法获得的临床靶点推进,大环肽非常适合在后 5 规则的制药环境中应对这些挑战。促进他们发现的是一系列高通量筛选方法,这些方法利用了大量基因编码化合物的随机库。这些技术受益于非天然部分的掺入,例如非蛋白原氨基酸或稳定的碳氢化合物主食。利用这些特征进行大环肽的战略结构设计有可能解决具有挑战性的靶标,例如蛋白质-蛋白质相互作用,这些靶标长期以来一直阻碍着研究工作。本文总结了用于发现大环肽的主要高通量技术的基本原理和最新进展,并重点介绍了它们靶向不可成药空间的具体部署。特别关注环肽环化和结构稳定的新设计指南和原则的开发,以及针对众所周知的难以接近的药物靶点取得的成功案例。
更新日期:2024-10-25
中文翻译:

为难以接近的药物靶标选择核苷酸编码的大环肽质量库
制药领域的技术进步和突破性发展正在以越来越坚定的坚持敲响“不可成药”堡垒的大门。值得注意的是,21 世纪出现了大环化合物,其中环肽特别令人感兴趣。这类新型的潜在候选药物占据了经典小分子药物和大型蛋白质疗法(如抗体)之间的巨大化学空间。随着研究朝着长期以来被认为无法获得的临床靶点推进,大环肽非常适合在后 5 规则的制药环境中应对这些挑战。促进他们发现的是一系列高通量筛选方法,这些方法利用了大量基因编码化合物的随机库。这些技术受益于非天然部分的掺入,例如非蛋白原氨基酸或稳定的碳氢化合物主食。利用这些特征进行大环肽的战略结构设计有可能解决具有挑战性的靶标,例如蛋白质-蛋白质相互作用,这些靶标长期以来一直阻碍着研究工作。本文总结了用于发现大环肽的主要高通量技术的基本原理和最新进展,并重点介绍了它们靶向不可成药空间的具体部署。特别关注环肽环化和结构稳定的新设计指南和原则的开发,以及针对众所周知的难以接近的药物靶点取得的成功案例。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号