当前位置:
X-MOL 学术
›
Cell Chem. Bio.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
RNA infrastructure profiling illuminates transcriptome structure in crowded spaces
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2024-10-23 , DOI: 10.1016/j.chembiol.2024.09.009 Lu Xiao, Linglan Fang, Wenrui Zhong, Eric T. Kool
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2024-10-23 , DOI: 10.1016/j.chembiol.2024.09.009 Lu Xiao, Linglan Fang, Wenrui Zhong, Eric T. Kool
|
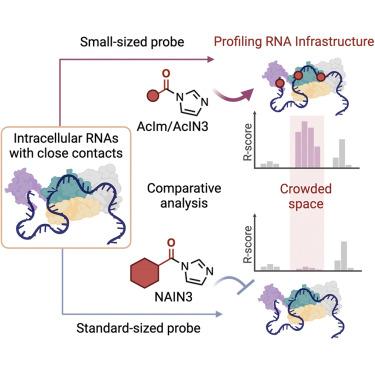
RNAs fold into compact structures and undergo protein interactions in cells. These occluded environments can block reagents that probe the underlying RNAs. Probes that can analyze structure in crowded settings can shed light on RNA biology. Here, we employ 2′-OH-reactive probes that are small enough to access folded RNA structure underlying close molecular contacts within cells, providing considerably broader coverage for intracellular RNA structural analysis. The data are analyzed first with well-characterized human ribosomal RNAs and then applied transcriptome-wide to polyadenylated transcripts. The smallest probe acetylimidazole (AcIm) yields 80% greater structural coverage than larger conventional reagent NAIN3, providing enhanced structural information in hundreds of transcripts. The acetyl probe also provides superior signals for identifying m6A modification sites in transcripts, particularly in sites that are inaccessible to a standard probe. Our strategy enables profiling RNA infrastructure, enhancing analysis of transcriptome structure, modification, and intracellular interactions, especially in spatially crowded settings.
中文翻译:

RNA 基础设施分析揭示了拥挤空间中的转录组结构
RNA 折叠成紧凑的结构并在细胞中发生蛋白质相互作用。这些封闭的环境会阻断探测底层 RNA 的试剂。可以在拥挤环境中分析结构的探针可以阐明 RNA 生物学。在这里,我们采用了 2′-OH 反应性探针,这些探针足够小,可以接触到细胞内紧密分子接触的折叠 RNA 结构,为细胞内 RNA 结构分析提供了相当广泛的覆盖范围。首先使用表征良好的人核糖体 RNA 分析数据,然后将转录组范围应用于多聚腺苷酸化转录本。最小的探针乙酰咪唑 (AcIm) 的结构覆盖度比较大的常规试剂 NAIN3 高 80%,可在数百个转录本中提供增强的结构信息。乙酰基探针还为识别转录本中的 m6A 修饰位点提供了出色的信号,特别是在标准探针无法接近的位点。我们的策略能够分析 RNA 基础设施,增强对转录组结构、修饰和细胞内相互作用的分析,尤其是在空间拥挤的环境中。
更新日期:2024-10-23
中文翻译:

RNA 基础设施分析揭示了拥挤空间中的转录组结构
RNA 折叠成紧凑的结构并在细胞中发生蛋白质相互作用。这些封闭的环境会阻断探测底层 RNA 的试剂。可以在拥挤环境中分析结构的探针可以阐明 RNA 生物学。在这里,我们采用了 2′-OH 反应性探针,这些探针足够小,可以接触到细胞内紧密分子接触的折叠 RNA 结构,为细胞内 RNA 结构分析提供了相当广泛的覆盖范围。首先使用表征良好的人核糖体 RNA 分析数据,然后将转录组范围应用于多聚腺苷酸化转录本。最小的探针乙酰咪唑 (AcIm) 的结构覆盖度比较大的常规试剂 NAIN3 高 80%,可在数百个转录本中提供增强的结构信息。乙酰基探针还为识别转录本中的 m6A 修饰位点提供了出色的信号,特别是在标准探针无法接近的位点。我们的策略能够分析 RNA 基础设施,增强对转录组结构、修饰和细胞内相互作用的分析,尤其是在空间拥挤的环境中。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号