当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Pre-training strategy for antiviral drug screening with low-data graph neural network: A case study in HIV-1 K103N reverse transcriptase
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-10-22 , DOI: 10.1002/jcc.27514 Kajjana Boonpalit, Hathaichanok Chuntakaruk, Jiramet Kinchagawat, Peter Wolschann, Supot Hannongbua, Thanyada Rungrotmongkol, Sarana Nutanong
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-10-22 , DOI: 10.1002/jcc.27514 Kajjana Boonpalit, Hathaichanok Chuntakaruk, Jiramet Kinchagawat, Peter Wolschann, Supot Hannongbua, Thanyada Rungrotmongkol, Sarana Nutanong
|
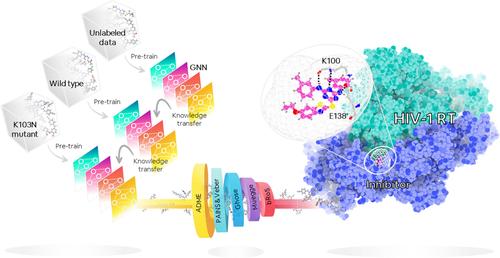
Graph neural networks (GNN) offer an alternative approach to boost the screening effectiveness in drug discovery. However, their efficacy is often hindered by limited datasets. To address this limitation, we introduced a robust GNN training framework, applied to various chemical databases to identify potent non-nucleoside reverse transcriptase inhibitors (NNRTIs) against the challenging K103N-mutated HIV-1 RT. Leveraging self-supervised learning (SSL) pre-training to tackle data scarcity, we screened 1,824,367 compounds, using multi-step approach that incorporated machine learning (ML)-based screening, analysis of absorption, distribution, metabolism, and excretion (ADME) prediction, drug-likeness properties, and molecular docking. Ultimately, 45 compounds were left as potential candidates with 17 of the compounds were previously identified as NNRTIs, exemplifying the model's efficacy. The remaining 28 compounds are anticipated to be repurposed for new uses. Molecular dynamics (MD) simulations on repurposed candidates unveiled two promising preclinical drugs: one designed against Plasmodium falciparum and the other serving as an antibacterial agent. Both have superior binding affinity compared to anti-HIV drugs. This conceptual framework could be adapted for other disease-specific therapeutics, facilitating the identification of potent compounds effective against both WT and mutants while revealing novel scaffolds for drug design and discovery.
中文翻译:

基于低数据图神经网络的抗病毒药物筛选预训练策略:以 HIV-1 K103N 逆转录酶为例
图神经网络 (GNN) 提供了一种提高药物发现筛选效果的替代方法。然而,它们的有效性往往受到有限数据集的阻碍。为了解决这一限制,我们引入了一个强大的 GNN 训练框架,应用于各种化学数据库,以识别针对具有挑战性的 K103N 突变 HIV-1 RT 的有效非核苷逆转录酶抑制剂 (NNRTI)。利用自我监督学习 (SSL) 预训练来解决数据稀缺问题,我们使用多步骤方法筛选了 1,824,367 种化合物,该方法结合了基于机器学习 (ML) 的筛选、吸收分析、 分布、代谢和排泄 (ADME) 预测、药物相似特性和分子对接。最终,留下了 45 种化合物作为潜在候选化合物,其中 17 种化合物之前被确定为 NNRTI,说明了该模型的功效。其余 28 种化合物预计将重新用于新用途。对重新利用的候选药物的分子动力学 (MD) 模拟揭示了两种有前途的临床前药物:一种是针对恶性疟原虫设计的,另一种用作抗菌剂。与抗 HIV 药物相比,两者都具有优异的结合亲和力。这个概念框架可以适用于其他疾病特异性疗法,有助于鉴定对 WT 和突变体有效的有效化合物,同时揭示用于药物设计和发现的新型支架。
更新日期:2024-10-22
中文翻译:

基于低数据图神经网络的抗病毒药物筛选预训练策略:以 HIV-1 K103N 逆转录酶为例
图神经网络 (GNN) 提供了一种提高药物发现筛选效果的替代方法。然而,它们的有效性往往受到有限数据集的阻碍。为了解决这一限制,我们引入了一个强大的 GNN 训练框架,应用于各种化学数据库,以识别针对具有挑战性的 K103N 突变 HIV-1 RT 的有效非核苷逆转录酶抑制剂 (NNRTI)。利用自我监督学习 (SSL) 预训练来解决数据稀缺问题,我们使用多步骤方法筛选了 1,824,367 种化合物,该方法结合了基于机器学习 (ML) 的筛选、吸收分析、 分布、代谢和排泄 (ADME) 预测、药物相似特性和分子对接。最终,留下了 45 种化合物作为潜在候选化合物,其中 17 种化合物之前被确定为 NNRTI,说明了该模型的功效。其余 28 种化合物预计将重新用于新用途。对重新利用的候选药物的分子动力学 (MD) 模拟揭示了两种有前途的临床前药物:一种是针对恶性疟原虫设计的,另一种用作抗菌剂。与抗 HIV 药物相比,两者都具有优异的结合亲和力。这个概念框架可以适用于其他疾病特异性疗法,有助于鉴定对 WT 和突变体有效的有效化合物,同时揭示用于药物设计和发现的新型支架。































 京公网安备 11010802027423号
京公网安备 11010802027423号