当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Multirelational Hypergraph Representation Learning for Predicting circRNA-miRNA Associations
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-21 , DOI: 10.1021/acs.jcim.4c01436 Wenjing Yin, Shudong Wang, Yuanyuan Zhang, Sibo Qiao, Wenhao Wu, Hengxiao Li
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-10-21 , DOI: 10.1021/acs.jcim.4c01436 Wenjing Yin, Shudong Wang, Yuanyuan Zhang, Sibo Qiao, Wenhao Wu, Hengxiao Li
|
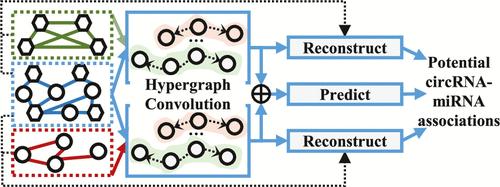
One of the principal functions of circular RNA (circRNA) is to participate in gene regulation by sponging microRNAs (miRNAs). Using accumulated circRNA-miRNA associations (CMAs) to construct computational models for predicting potential associations provides a crucial tool for accelerating the validation of reliable associations through traditional experiments. Nevertheless, the current prediction models are constrained in their capacity to represent the higher-order relationships of CMAs and thus require further enhancement in terms of their predictive efficacy. In order to address this issue, we propose a new model based on multirelational hypergraph representation learning (MRHRL). This model employs hypergraphs to capture various higher-order relationships among RNAs and aggregates complementary information through a view attention mechanism. Furthermore, MRHRL introduces a hyperedge-level reconstruction task, jointly optimizing the prediction and reconstruction tasks within a unified framework to uncover potential information, thereby enhancing the model’s predictive and generalization capabilities. Experiments conducted on three real-world data sets demonstrate that MRHRL achieves satisfactory results in CMAs prediction, significantly outperforming existing prediction models.
中文翻译:

用于预测 circRNA-miRNA 关联的多关系超图表示学习
环状 RNA (circRNA) 的主要功能之一是通过海绵 microRNA (miRNA) 参与基因调控。使用累积的 circRNA-miRNA 关联 (CMA) 构建用于预测潜在关联的计算模型,为通过传统实验加速验证可靠关联提供了重要工具。然而,目前的预测模型在表示 CMA 的高阶关系的能力方面受到限制,因此需要进一步增强其预测效能。为了解决这个问题,我们提出了一种基于多关系超图表示学习 (MRHRL) 的新模型。该模型采用超图来捕获 RNA 之间的各种高阶关系,并通过视图注意力机制聚合互补信息。此外,MRHRL 引入了超边缘级别的重建任务,在统一的框架内共同优化预测和重建任务,以发现潜在信息,从而增强模型的预测和泛化能力。在三个真实数据集上进行的实验表明,MRHRL 在 CMA 预测方面取得了令人满意的结果,明显优于现有的预测模型。
更新日期:2024-10-21
中文翻译:

用于预测 circRNA-miRNA 关联的多关系超图表示学习
环状 RNA (circRNA) 的主要功能之一是通过海绵 microRNA (miRNA) 参与基因调控。使用累积的 circRNA-miRNA 关联 (CMA) 构建用于预测潜在关联的计算模型,为通过传统实验加速验证可靠关联提供了重要工具。然而,目前的预测模型在表示 CMA 的高阶关系的能力方面受到限制,因此需要进一步增强其预测效能。为了解决这个问题,我们提出了一种基于多关系超图表示学习 (MRHRL) 的新模型。该模型采用超图来捕获 RNA 之间的各种高阶关系,并通过视图注意力机制聚合互补信息。此外,MRHRL 引入了超边缘级别的重建任务,在统一的框架内共同优化预测和重建任务,以发现潜在信息,从而增强模型的预测和泛化能力。在三个真实数据集上进行的实验表明,MRHRL 在 CMA 预测方面取得了令人满意的结果,明显优于现有的预测模型。

































 京公网安备 11010802027423号
京公网安备 11010802027423号