当前位置:
X-MOL 学术
›
Biotechnol. Bioeng.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Transfer learning Bayesian optimization for competitor DNA molecule design for use in diagnostic assays
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2024-10-16 , DOI: 10.1002/bit.28854 Ruby Sedgwick, John P. Goertz, Molly M. Stevens, Ruth Misener, Mark van der Wilk
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2024-10-16 , DOI: 10.1002/bit.28854 Ruby Sedgwick, John P. Goertz, Molly M. Stevens, Ruth Misener, Mark van der Wilk
|
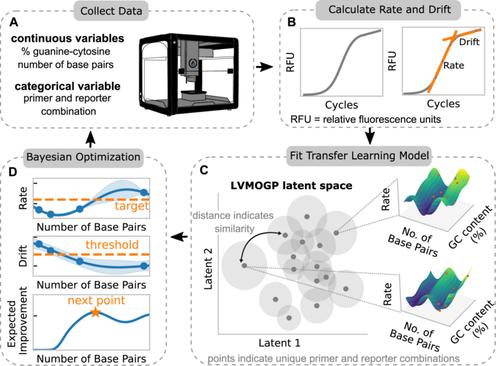
With the rise in engineered biomolecular devices, there is an increased need for tailor‐made biological sequences. Often, many similar biological sequences need to be made for a specific application meaning numerous, sometimes prohibitively expensive, lab experiments are necessary for their optimization. This paper presents a transfer learning design of experiments workflow to make this development feasible. By combining a transfer learning surrogate model with Bayesian optimization, we show how the total number of experiments can be reduced by sharing information between optimization tasks. We demonstrate the reduction in the number of experiments using data from the development of DNA competitors for use in an amplification‐based diagnostic assay. We use cross‐validation to compare the predictive accuracy of different transfer learning models, and then compare the performance of the models for both single objective and penalized optimization tasks.
中文翻译:

用于诊断分析的竞争对手 DNA 分子设计的迁移学习贝叶斯优化
随着工程生物分子装置的兴起,对定制生物序列的需求也越来越大。通常,需要为特定应用制作许多类似的生物序列,这意味着需要进行大量有时非常昂贵的实验室实验来优化它们。本文提出了一种实验的迁移学习设计工作流程,以使这种开发可行。通过将迁移学习代理模型与贝叶斯优化相结合,我们展示了如何通过在优化任务之间共享信息来减少实验总数。我们使用 DNA 竞争对手开发的数据证明实验数量的减少,用于基于扩增的诊断分析。我们使用交叉验证来比较不同迁移学习模型的预测准确性,然后比较模型在单目标和惩罚优化任务中的性能。
更新日期:2024-10-16
中文翻译:

用于诊断分析的竞争对手 DNA 分子设计的迁移学习贝叶斯优化
随着工程生物分子装置的兴起,对定制生物序列的需求也越来越大。通常,需要为特定应用制作许多类似的生物序列,这意味着需要进行大量有时非常昂贵的实验室实验来优化它们。本文提出了一种实验的迁移学习设计工作流程,以使这种开发可行。通过将迁移学习代理模型与贝叶斯优化相结合,我们展示了如何通过在优化任务之间共享信息来减少实验总数。我们使用 DNA 竞争对手开发的数据证明实验数量的减少,用于基于扩增的诊断分析。我们使用交叉验证来比较不同迁移学习模型的预测准确性,然后比较模型在单目标和惩罚优化任务中的性能。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号