Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Probing the role of ligation and exonuclease digestion towards non-specific amplification in bioanalytical RCA assays
Analyst ( IF 3.6 ) Pub Date : 2024-10-10 , DOI: 10.1039/d4an00866a Vandana Kuttappan Nair, Chandrika Sharma, Shrawan Kumar, Mrittika Sengupta, Souradyuti Ghosh
Analyst ( IF 3.6 ) Pub Date : 2024-10-10 , DOI: 10.1039/d4an00866a Vandana Kuttappan Nair, Chandrika Sharma, Shrawan Kumar, Mrittika Sengupta, Souradyuti Ghosh
|
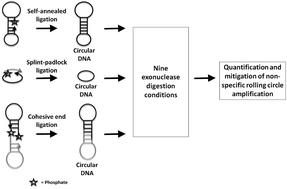
Non-specific amplification (NSA, amplification in the absence of a target analyte) in bioanalytical rolling circle amplification (RCA) assays, especially those involving pre-synthesized circular DNA (cDNA), affects its analytical sensitivity. Despite extensive development of RCA-based bioanalytical methods, the NSA in RCA remains uncharacterized in terms of its magnitude or origin. NSA may originate from inefficient ligation or succeeding cDNA purification steps. This study comprehensively quantifies NSA across several ligation and digestion techniques for the first time since the innovation of RCA. To quantify the NSA in RCA, cDNAs were prepared using self-annealing, splint-padlock, or cohesive end ligations. The cDNAs were then subjected to nine different exonuclease digestion steps and quantified for NSA under linear as well as hyperbranched RCA conditions. We investigated buffer compositions, divalent ion concentrations, single or dual enzyme digestion, cohesive end lengths, and splint lengths. The optimized conditions successfully mitigated absolute NSA by 30–100-fold and relative NSA (normalized against primer-assisted RCA) to ∼5%. Besides understanding the mechanistic origin of NSA, novel aspects of enzyme–substrate selectivity, buffer composition, and the role of divalent ions were discovered. With increasing bioanalytical RCA applications, this study will help standardize NSA-free assays.
中文翻译:

探索连接和核酸外切酶消化对生物分析 RCA 分析中非特异性扩增的作用
生物分析滚环扩增 (RCA) 分析中的非特异性扩增(NSA,在没有目标分析物的情况下进行扩增),尤其是涉及预合成环状 DNA (cDNA) 的分析,会影响其分析灵敏度。尽管基于 RCA 的生物分析方法得到了广泛发展,但 RCA 中的 NSA 在其大小或来源方面仍未得到表征。NSA 可能源于低效的连接或后续的 cDNA 纯化步骤。自 RCA 创新以来,本研究首次全面量化了几种连接和消化技术的 NSA。为了定量 RCA 中的 NSA,使用自退火、夹板挂锁或粘性末端连接制备 cDNA。然后对 cDNA 进行 9 个不同的核酸外切酶消化步骤,并在线性和超支化 RCA 条件下对 NSA 进行定量。我们研究了缓冲液组成、二价离子浓度、单酶或双酶消化、内聚末端长度和夹板长度。优化条件成功地将绝对 NSA 降低了 30-100 倍,将相对 NSA(针对引物辅助 RCA 标准化)降低到 ∼5%。除了了解 NSA 的机制起源外,还发现了酶-底物选择性、缓冲液组成和二价离子作用的新方面。随着生物分析 RCA 应用的增加,本研究将有助于标准化无 NSA 检测。
更新日期:2024-10-10
中文翻译:

探索连接和核酸外切酶消化对生物分析 RCA 分析中非特异性扩增的作用
生物分析滚环扩增 (RCA) 分析中的非特异性扩增(NSA,在没有目标分析物的情况下进行扩增),尤其是涉及预合成环状 DNA (cDNA) 的分析,会影响其分析灵敏度。尽管基于 RCA 的生物分析方法得到了广泛发展,但 RCA 中的 NSA 在其大小或来源方面仍未得到表征。NSA 可能源于低效的连接或后续的 cDNA 纯化步骤。自 RCA 创新以来,本研究首次全面量化了几种连接和消化技术的 NSA。为了定量 RCA 中的 NSA,使用自退火、夹板挂锁或粘性末端连接制备 cDNA。然后对 cDNA 进行 9 个不同的核酸外切酶消化步骤,并在线性和超支化 RCA 条件下对 NSA 进行定量。我们研究了缓冲液组成、二价离子浓度、单酶或双酶消化、内聚末端长度和夹板长度。优化条件成功地将绝对 NSA 降低了 30-100 倍,将相对 NSA(针对引物辅助 RCA 标准化)降低到 ∼5%。除了了解 NSA 的机制起源外,还发现了酶-底物选择性、缓冲液组成和二价离子作用的新方面。随着生物分析 RCA 应用的增加,本研究将有助于标准化无 NSA 检测。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号