当前位置:
X-MOL 学术
›
Mol. Nutr. Food Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Use of FISH‐FLOW as a Method for the Identification and Quantification of Bacterial Populations
Molecular Nutrition & Food Research ( IF 4.5 ) Pub Date : 2024-10-04 , DOI: 10.1002/mnfr.202400494 Jorge Enrique Vazquez Bucheli, Yuri Lee, Bobae Kim, Nuno F. Azevedo, Andreia S. Azevedo, Svetoslav Dimitrov Todorov, Yosep Ji, Hyeji Kang, Wilhelm H. Holzapfel
Molecular Nutrition & Food Research ( IF 4.5 ) Pub Date : 2024-10-04 , DOI: 10.1002/mnfr.202400494 Jorge Enrique Vazquez Bucheli, Yuri Lee, Bobae Kim, Nuno F. Azevedo, Andreia S. Azevedo, Svetoslav Dimitrov Todorov, Yosep Ji, Hyeji Kang, Wilhelm H. Holzapfel
|
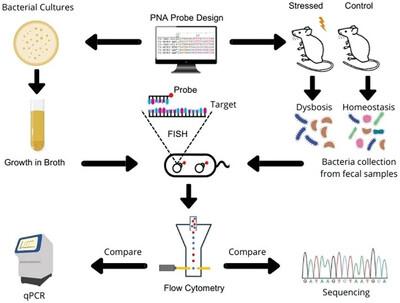
The gastrointestinal tract (GIT) harbors the largest group of microbiotas among the microbial communities of the human host. The resident organisms typical of a healthy gut are well adapted to the gastrointestinal environment while alteration of these populations can trigger disorders that may affect the health and well‐being of the host. Various investigations have applied different tools to study bacterial communities in the gut and their correlation with gastrointestinal disorders such as inflammatory bowel disease (IBD), obesity, and diabetes. This study proposes fluorescent in situ hybridization, combined with flow cytometry (FISH‐FLOW), as an alternative approach for phylum level identification of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria and quantification of target bacteria from the GIT based on analysis of fecal samples, where results are validated by quantitative polymerase chain reaction (qPCR) and 16S ribosomal ribonucleic acid (16s rRNA) sequencing. The results obtained via FISH‐FLOW experimental approach show high specificity for the developed probes for hybridization with the target bacteria. The study, therefore, suggests the FISH‐FLOW as a reliable method for studying bacterial communities in the gut with results correlating well with those of metagenomic investigations of the same fecal samples.
中文翻译:

使用 FISH-FLOW 作为细菌种群鉴定和定量的方法
胃肠道 (GIT) 在人类宿主的微生物群落中拥有最大的微生物群。健康肠道的典型常驻生物体很好地适应了胃肠道环境,而这些种群的改变会引发可能影响宿主健康和福祉的疾病。各种研究应用了不同的工具来研究肠道中的细菌群落及其与胃肠道疾病(如炎症性肠病 (IBD)、肥胖和糖尿病)的相关性。本研究提出了荧光原位杂交,结合流式细胞术 (FISH-FLOW),作为门水平鉴定厚壁菌门、拟杆菌门、放线菌门和变形菌门的替代方法,以及基于粪便样本分析的 GIT 中靶细菌的定量,其中结果通过定量聚合酶链反应 (qPCR) 和 16S 核糖体核糖核酸 (16S rRNA) 测序进行验证。通过 FISH-FLOW 实验方法获得的结果显示,开发的探针与目标细菌杂交具有高度特异性。因此,该研究表明 FISH-FLOW 是研究肠道细菌群落的可靠方法,其结果与相同粪便样本的宏基因组研究结果非常相关。
更新日期:2024-10-04
中文翻译:

使用 FISH-FLOW 作为细菌种群鉴定和定量的方法
胃肠道 (GIT) 在人类宿主的微生物群落中拥有最大的微生物群。健康肠道的典型常驻生物体很好地适应了胃肠道环境,而这些种群的改变会引发可能影响宿主健康和福祉的疾病。各种研究应用了不同的工具来研究肠道中的细菌群落及其与胃肠道疾病(如炎症性肠病 (IBD)、肥胖和糖尿病)的相关性。本研究提出了荧光原位杂交,结合流式细胞术 (FISH-FLOW),作为门水平鉴定厚壁菌门、拟杆菌门、放线菌门和变形菌门的替代方法,以及基于粪便样本分析的 GIT 中靶细菌的定量,其中结果通过定量聚合酶链反应 (qPCR) 和 16S 核糖体核糖核酸 (16S rRNA) 测序进行验证。通过 FISH-FLOW 实验方法获得的结果显示,开发的探针与目标细菌杂交具有高度特异性。因此,该研究表明 FISH-FLOW 是研究肠道细菌群落的可靠方法,其结果与相同粪便样本的宏基因组研究结果非常相关。


















































 京公网安备 11010802027423号
京公网安备 11010802027423号