当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Development of a machine learning-based target-specific scoring function for structure-based binding affinity prediction for human dihydroorotate dehydrogenase inhibitors
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-09-26 , DOI: 10.1002/jcc.27510 Jinhui Meng, Li Zhang, Zhe He, Mengfeng Hu, Jinhan Liu, Wenzhuo Bao, Qifeng Tian, Huawei Feng, Hongsheng Liu
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-09-26 , DOI: 10.1002/jcc.27510 Jinhui Meng, Li Zhang, Zhe He, Mengfeng Hu, Jinhan Liu, Wenzhuo Bao, Qifeng Tian, Huawei Feng, Hongsheng Liu
|
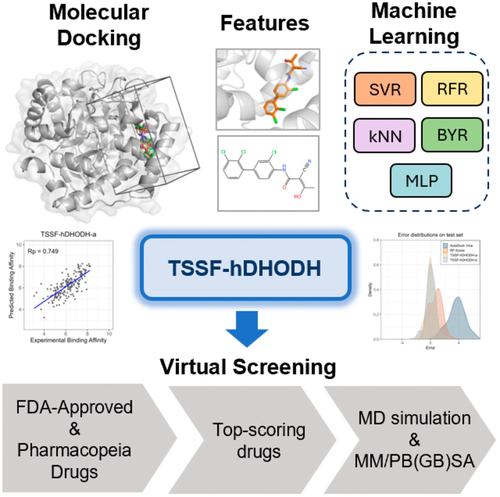
Human dihydroorotate dehydrogenase (hDHODH) is a flavin mononucleotide-dependent enzyme that can limit de novo pyrimidine synthesis, making it a therapeutic target for diseases such as autoimmune disorders and cancer. In this study, using the docking structures of complexes generated by AutoDock Vina, we integrate interaction features and ligand features, and employ support vector regression to develop a target-specific scoring function for hDHODH (TSSF-hDHODH). The Pearson correlation coefficient values of TSSF-hDHODH in the cross-validation and external validation are 0.86 and 0.74, respectively, both of which are far superior to those of classic scoring function AutoDock Vina and random forest (RF) based generic scoring function RF-Score. TSSF-hDHODH is further used for the virtual screening of potential inhibitors in the FDA-Approved & Pharmacopeia Drug Library. In conjunction with the results from molecular dynamics simulations, crizotinib is identified as a candidate for subsequent structural optimization. This study can be useful for the discovery of hDHODH inhibitors and the development of scoring functions for additional targets.
中文翻译:

开发基于机器学习的靶标特异性评分函数,用于人二氢乳清酸脱氢酶抑制剂的基于结构的结合亲和力预测
人二氢乳清酸脱氢酶 (hDHODH) 是一种黄素单核苷酸依赖性酶,可限制嘧啶从头合成,使其成为自身免疫性疾病和癌症等疾病的治疗靶点。在这项研究中,利用 AutoDock Vina 生成的复合物的对接结构,我们整合了相互作用特征和配体特征,并采用支持向量回归开发了 hDHODH 的目标特异性评分函数 (TSSF-hDHODH)。TSSF-hDHODH 在交叉验证和外部验证中的 Pearson 相关系数值分别为 0.86 和 0.74,均远优于经典评分函数 AutoDock Vina 和基于随机森林 (RF) 的通用评分函数 RF-Score。TSSF-hDHODH进一步用于在FDA批准和药典药物库中虚拟筛选潜在抑制剂。结合分子动力学模拟的结果,克唑替尼被确定为后续结构优化的候选药物。这项研究可用于发现 hDHODH 抑制剂和开发其他靶点的评分函数。
更新日期:2024-09-26
中文翻译:

开发基于机器学习的靶标特异性评分函数,用于人二氢乳清酸脱氢酶抑制剂的基于结构的结合亲和力预测
人二氢乳清酸脱氢酶 (hDHODH) 是一种黄素单核苷酸依赖性酶,可限制嘧啶从头合成,使其成为自身免疫性疾病和癌症等疾病的治疗靶点。在这项研究中,利用 AutoDock Vina 生成的复合物的对接结构,我们整合了相互作用特征和配体特征,并采用支持向量回归开发了 hDHODH 的目标特异性评分函数 (TSSF-hDHODH)。TSSF-hDHODH 在交叉验证和外部验证中的 Pearson 相关系数值分别为 0.86 和 0.74,均远优于经典评分函数 AutoDock Vina 和基于随机森林 (RF) 的通用评分函数 RF-Score。TSSF-hDHODH进一步用于在FDA批准和药典药物库中虚拟筛选潜在抑制剂。结合分子动力学模拟的结果,克唑替尼被确定为后续结构优化的候选药物。这项研究可用于发现 hDHODH 抑制剂和开发其他靶点的评分函数。

































 京公网安备 11010802027423号
京公网安备 11010802027423号