当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
RNAfcg: RNA Flexibility Prediction Based on Topological Centrality and Global Features
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-09-14 , DOI: 10.1021/acs.jcim.4c00848 Fubin Chang 1 , Lamei Liu 1 , Fangrui Hu 1 , Xiaohan Sun 1 , Yingchun Zhao 1 , Na Zhang 1 , Chunhua Li 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-09-14 , DOI: 10.1021/acs.jcim.4c00848 Fubin Chang 1 , Lamei Liu 1 , Fangrui Hu 1 , Xiaohan Sun 1 , Yingchun Zhao 1 , Na Zhang 1 , Chunhua Li 1
Affiliation
|
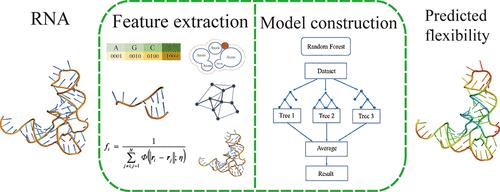
The dynamics of RNAs are related intimately to their functions. Molecular flexibility, as a starting point for understanding their dynamics, has been utilized to predict many characteristics associated with their functions. Since the experimental measurement methods are time-consuming and labor-intensive, it is urgently needed to develop reliable theoretical methods to predict RNA flexibility. In this work, we develop an effective machine learning method, RNAfcg, to predict RNA flexibility, where the Random Forest (RF) is trained by features including the topological centralities, flexibility–rigidity index, and global characteristics first introduced by us, as well as some traditional sequence and structural features. The analyses show that the three types of features introduced first have significant contributions to RNA flexibility prediction, among which the topological type contributes the most, which indicates the importance of structural topology in determining RNA flexibility. The performance comparison indicates that RNAfcg outperforms the state-of-the-art machine learning methods and the commonly used Gaussian Network Model (GNM) models, achieving a much higher Pearson correlation coefficient (PCC) of 0.6619 on the test data set. This work is helpful for understanding RNA dynamics and can be used to predict RNA function information. The source code is available at https://github.com/ChunhuaLab/RNAfcg/.
中文翻译:

RNAfcg:基于拓扑中心性和全局特征的 RNA 灵活性预测
RNA 的动力学与其功能密切相关。分子灵活性作为理解其动力学的起点,已被用于预测与其功能相关的许多特征。由于实验测量方法耗时且劳动密集,因此迫切需要开发可靠的理论方法来预测 RNA 灵活性。在这项工作中,我们开发了一种有效的机器学习方法 RNAfcg 来预测 RNA 灵活性,其中随机森林 (RF) 由我们首先引入的拓扑中心性、灵活性-刚性指数和全局特征以及一些传统的序列和结构特征等特征进行训练。分析表明,首先引入的 3 类特征对 RNA 灵活性预测有显著贡献,其中拓扑类型贡献最大,这表明结构拓扑结构在确定 RNA 灵活性方面的重要性。性能比较表明,RNAfcg 的性能优于最先进的机器学习方法和常用的高斯网络模型 (GNM) 模型,在测试数据集上实现了 0.6619 的更高皮尔逊相关系数 (PCC)。这项工作有助于理解 RNA 动力学,可用于预测 RNA 功能信息。源代码可在 https://github.com/ChunhuaLab/RNAfcg/ 获取。
更新日期:2024-09-19
中文翻译:

RNAfcg:基于拓扑中心性和全局特征的 RNA 灵活性预测
RNA 的动力学与其功能密切相关。分子灵活性作为理解其动力学的起点,已被用于预测与其功能相关的许多特征。由于实验测量方法耗时且劳动密集,因此迫切需要开发可靠的理论方法来预测 RNA 灵活性。在这项工作中,我们开发了一种有效的机器学习方法 RNAfcg 来预测 RNA 灵活性,其中随机森林 (RF) 由我们首先引入的拓扑中心性、灵活性-刚性指数和全局特征以及一些传统的序列和结构特征等特征进行训练。分析表明,首先引入的 3 类特征对 RNA 灵活性预测有显著贡献,其中拓扑类型贡献最大,这表明结构拓扑结构在确定 RNA 灵活性方面的重要性。性能比较表明,RNAfcg 的性能优于最先进的机器学习方法和常用的高斯网络模型 (GNM) 模型,在测试数据集上实现了 0.6619 的更高皮尔逊相关系数 (PCC)。这项工作有助于理解 RNA 动力学,可用于预测 RNA 功能信息。源代码可在 https://github.com/ChunhuaLab/RNAfcg/ 获取。































 京公网安备 11010802027423号
京公网安备 11010802027423号