当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
MMFA-DTA: Multimodal Feature Attention Fusion Network for Drug-Target Affinity Prediction for Drug Repurposing Against SARS-CoV-2
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2024-09-13 , DOI: 10.1021/acs.jctc.4c00663 Guanxing Chen, Haohuai He, Qiujie Lv, Lu Zhao, Calvin Yu-Chian Chen
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2024-09-13 , DOI: 10.1021/acs.jctc.4c00663 Guanxing Chen, Haohuai He, Qiujie Lv, Lu Zhao, Calvin Yu-Chian Chen
|
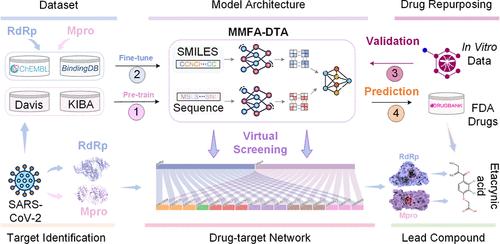
The continuous emergence of novel infectious diseases poses a significant threat to global public health security, necessitating the development of small-molecule inhibitors that directly target pathogens. The RNA-dependent RNA polymerase (RdRp) and main protease (Mpro) of SARS-CoV-2 have been validated as potential key antiviral drug targets for the treatment of COVID-19. However, the conventional new drug R&D cycle takes 10–15 years, failing to meet the urgent needs during epidemics. Here, we propose a general multimodal deep learning framework for drug repurposing, MMFA-DTA, to enable rapid virtual screening of known drugs and significantly improve discovery efficiency. By extracting graph topological and sequence features from both small molecules and proteins, we design attention mechanisms to achieve dynamic fusion across modalities. Results demonstrate the superior performance of MMFA-DTA in drug-target affinity prediction over several state-of-the-art baseline methods on Davis and KIBA data sets, validating the benefits of heterogeneous information integration for representation learning and interaction modeling. Further fine-tuning on COVID-19-relevant bioactivity data enhances model predictions for critical SARS-CoV-2 enzymes. Case studies screening the FDA-approved drug library successfully identify etacrynic acid as the potential lead compound against both RdRp and Mpro. Molecular dynamics simulations further confirm the stability and binding affinity of etacrynic acid to these targets. This study proves the great potential and advantages of deep learning and drug repurposing strategies in supporting antiviral drug discovery. The proposed general and rapid response computational framework holds significance for preparedness against future public health events.
中文翻译:

MMFA-DTA:用于药物靶点亲和力预测的多模态特征注意融合网络,用于针对 SARS-CoV-2 的药物再利用
新型传染病的不断出现对全球公共卫生安全构成重大威胁,需要开发直接针对病原体的小分子抑制剂。 SARS-CoV-2 的 RNA 依赖性 RNA 聚合酶 (RdRp) 和主要蛋白酶 (Mpro) 已被验证为治疗 COVID-19 的潜在关键抗病毒药物靶点。然而,传统的新药研发周期需要10-15年,无法满足疫情期间的迫切需求。在这里,我们提出了一种用于药物再利用的通用多模态深度学习框架 MMFA-DTA,以实现已知药物的快速虚拟筛选并显着提高发现效率。通过从小分子和蛋白质中提取图拓扑和序列特征,我们设计了注意力机制来实现跨模态的动态融合。结果表明,在 Davis 和 KIBA 数据集上,MMFA-DTA 在药物靶点亲和力预测方面优于几种最先进的基线方法,验证了异构信息集成对于表示学习和交互建模的好处。对 COVID-19 相关生物活性数据的进一步微调可增强对关键 SARS-CoV-2 酶的模型预测。筛选 FDA 批准的药物库的案例研究成功地将依他尼酸确定为对抗 RdRp 和 Mpro 的潜在先导化合物。分子动力学模拟进一步证实了依他尼酸与这些靶标的稳定性和结合亲和力。这项研究证明了深度学习和药物再利用策略在支持抗病毒药物发现方面的巨大潜力和优势。 所提出的通用和快速响应计算框架对于应对未来公共卫生事件具有重要意义。
更新日期:2024-09-13
中文翻译:

MMFA-DTA:用于药物靶点亲和力预测的多模态特征注意融合网络,用于针对 SARS-CoV-2 的药物再利用
新型传染病的不断出现对全球公共卫生安全构成重大威胁,需要开发直接针对病原体的小分子抑制剂。 SARS-CoV-2 的 RNA 依赖性 RNA 聚合酶 (RdRp) 和主要蛋白酶 (Mpro) 已被验证为治疗 COVID-19 的潜在关键抗病毒药物靶点。然而,传统的新药研发周期需要10-15年,无法满足疫情期间的迫切需求。在这里,我们提出了一种用于药物再利用的通用多模态深度学习框架 MMFA-DTA,以实现已知药物的快速虚拟筛选并显着提高发现效率。通过从小分子和蛋白质中提取图拓扑和序列特征,我们设计了注意力机制来实现跨模态的动态融合。结果表明,在 Davis 和 KIBA 数据集上,MMFA-DTA 在药物靶点亲和力预测方面优于几种最先进的基线方法,验证了异构信息集成对于表示学习和交互建模的好处。对 COVID-19 相关生物活性数据的进一步微调可增强对关键 SARS-CoV-2 酶的模型预测。筛选 FDA 批准的药物库的案例研究成功地将依他尼酸确定为对抗 RdRp 和 Mpro 的潜在先导化合物。分子动力学模拟进一步证实了依他尼酸与这些靶标的稳定性和结合亲和力。这项研究证明了深度学习和药物再利用策略在支持抗病毒药物发现方面的巨大潜力和优势。 所提出的通用和快速响应计算框架对于应对未来公共卫生事件具有重要意义。































 京公网安备 11010802027423号
京公网安备 11010802027423号