当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Discovery of megapolipeptins by genome mining of a Burkholderiales bacteria collection
Chemical Science ( IF 7.6 ) Pub Date : 2024-09-13 , DOI: 10.1039/d4sc03594a Bruno S. Paulo, Michael J. J. Recchia, Sanghoon Lee, Claire H. Fergusson, Sean B. Romanowski, Antonio Hernandez, Nyssa Krull, Dennis Y. Liu, Hannah Cavanagh, Allyson Bos, Christopher A. Gray, Brian T. Murphy, Roger G. Linington, Alessandra S. Eustaquio
Chemical Science ( IF 7.6 ) Pub Date : 2024-09-13 , DOI: 10.1039/d4sc03594a Bruno S. Paulo, Michael J. J. Recchia, Sanghoon Lee, Claire H. Fergusson, Sean B. Romanowski, Antonio Hernandez, Nyssa Krull, Dennis Y. Liu, Hannah Cavanagh, Allyson Bos, Christopher A. Gray, Brian T. Murphy, Roger G. Linington, Alessandra S. Eustaquio
|
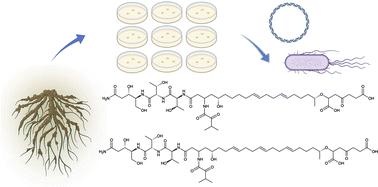
Burkholderiales bacteria have emerged as a promising source of structurally diverse natural products that are expected to play important ecological and industrial roles. This order ranks in the top three in terms of predicted natural product diversity from available genomes, warranting further genome sequencing efforts. However, a major hurdle in obtaining the predicted products is that biosynthetic genes are often ‘silent’ or poorly expressed. Here we report complementary strain isolation, genomics, metabolomics, and synthetic biology approaches to enable natural product discovery. First, we built a collection of 316 rhizosphere-derived Burkholderiales strains over the course of five years. We then selected 115 strains for sequencing using the mass spectrometry pipeline IDBac to avoid strain redundancy. After predicting and comparing the biosynthetic potential of each strain, a biosynthetic gene cluster that was silent in the native Paraburkholderia megapolitana and Paraburkholderia acidicola producers was cloned and activated by heterologous expression in a Burkholderia sp. host, yielding megapolipeptins A and B. Megapolipeptins are unusual polyketide, nonribosomal peptide, and polyunsaturated fatty acid hybrids that show low structural similarity to known natural products, highlighting the advantage of our Burkholderiales genomics-driven and synthetic biology-enabled pipeline to discover novel natural products.
中文翻译:

通过对 Burkholderiales 细菌集合进行基因组挖掘发现 megapolipeptins
Burkholderiales 细菌已成为结构多样的天然产物的有前途的来源,有望发挥重要的生态和工业作用。就可用基因组预测的天然产物多样性而言,该顺序排名前三,需要进一步的基因组测序工作。然而,获得预测产物的一个主要障碍是生物合成基因通常是“沉默的”或表达不佳的。在这里,我们报告了互补的菌株分离、基因组学、代谢组学和合成生物学方法,以实现天然产物的发现。首先,我们在五年内建立了 316 种根际衍生的 Burkholderiales 菌株的集合。然后,我们选择了 115 个菌株使用质谱管道 IDBac 进行测序,以避免菌株冗余。在预测和比较每种菌株的生物合成潜力后,克隆了一个在天然 Paraburkholderia megapolitana 和 Paraburkholderia acidicola 生产者中沉默的生物合成基因簇,并通过在伯克霍尔德菌属宿主中的异源表达激活,产生 megapolipeptins A 和 B。Megapolipeptins 是不寻常的聚酮、非核糖体肽和多不饱和脂肪酸杂交体,与已知的结构相似性较低天然产物,突出了我们的 Burkholderiales 基因组学驱动和合成生物学支持管道在发现新型天然产物方面的优势。
更新日期:2024-09-13
中文翻译:

通过对 Burkholderiales 细菌集合进行基因组挖掘发现 megapolipeptins
Burkholderiales 细菌已成为结构多样的天然产物的有前途的来源,有望发挥重要的生态和工业作用。就可用基因组预测的天然产物多样性而言,该顺序排名前三,需要进一步的基因组测序工作。然而,获得预测产物的一个主要障碍是生物合成基因通常是“沉默的”或表达不佳的。在这里,我们报告了互补的菌株分离、基因组学、代谢组学和合成生物学方法,以实现天然产物的发现。首先,我们在五年内建立了 316 种根际衍生的 Burkholderiales 菌株的集合。然后,我们选择了 115 个菌株使用质谱管道 IDBac 进行测序,以避免菌株冗余。在预测和比较每种菌株的生物合成潜力后,克隆了一个在天然 Paraburkholderia megapolitana 和 Paraburkholderia acidicola 生产者中沉默的生物合成基因簇,并通过在伯克霍尔德菌属宿主中的异源表达激活,产生 megapolipeptins A 和 B。Megapolipeptins 是不寻常的聚酮、非核糖体肽和多不饱和脂肪酸杂交体,与已知的结构相似性较低天然产物,突出了我们的 Burkholderiales 基因组学驱动和合成生物学支持管道在发现新型天然产物方面的优势。































 京公网安备 11010802027423号
京公网安备 11010802027423号