当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Decomposition of Forces in Protein: Methodology and General Properties
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-09-11 , DOI: 10.1021/acs.jcim.4c00716 Pengbo Song 1 , Qiaojing Huang 1 , Wenyu Li 1 , Maodong Li 2 , Zhirong Liu 1, 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-09-11 , DOI: 10.1021/acs.jcim.4c00716 Pengbo Song 1 , Qiaojing Huang 1 , Wenyu Li 1 , Maodong Li 2 , Zhirong Liu 1, 3
Affiliation
|
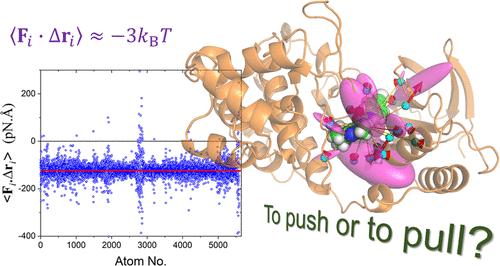
In contrast to the central role played by the structure of biomolecules, the complementary force-based view has received little attention in past studies. Here, we proposed a simple method for the force decomposition of multibody interactions and provided some techniques to analyze and visualize the general behavior of forces in proteins. It was shown that atomic forces fluctuate at a magnitude of about 3000 pN, which is huge in the context of cell biology. Remarkably, the average scalar product between atomic force and displacement universally approximates −3kBT. This is smaller by an order of magnitude than the simple product of their fluctuation magnitudes due to the unexpectedly weak correlation between the directions of force and displacement. The pairwise forces are highly anisotropic, with elongated fluctuation ellipsoids. Residue–residue forces can be attractive or repulsive (despite being more likely to be attractive), forming some kind of tensegrity structure stabilized by a complicated network of forces. Being able to understand and predict the interaction network provides a basis for rational drug design and uncovering molecular recognition mechanisms.
中文翻译:

蛋白质中力的分解:方法和一般性质
与生物分子结构所发挥的核心作用相反,基于互补力的观点在过去的研究中很少受到关注。在这里,我们提出了一种简单的多体相互作用力分解方法,并提供了一些技术来分析和可视化蛋白质中力的一般行为。结果表明,原子力的波动幅度约为 3000 pN,这对于细胞生物学来说是巨大的。值得注意的是,原子力和位移之间的平均标量积普遍接近 -3 k B T 。由于力和位移方向之间的相关性出乎意料地微弱,这比它们波动幅度的简单乘积小一个数量级。成对力具有高度各向异性,具有拉长的涨落椭球体。残留力-残留力可以是吸引力或排斥力(尽管更有可能具有吸引力),形成某种由复杂的力网络稳定的张拉整体结构。能够理解和预测相互作用网络为合理的药物设计和揭示分子识别机制提供了基础。
更新日期:2024-09-11
中文翻译:

蛋白质中力的分解:方法和一般性质
与生物分子结构所发挥的核心作用相反,基于互补力的观点在过去的研究中很少受到关注。在这里,我们提出了一种简单的多体相互作用力分解方法,并提供了一些技术来分析和可视化蛋白质中力的一般行为。结果表明,原子力的波动幅度约为 3000 pN,这对于细胞生物学来说是巨大的。值得注意的是,原子力和位移之间的平均标量积普遍接近 -3 k B T 。由于力和位移方向之间的相关性出乎意料地微弱,这比它们波动幅度的简单乘积小一个数量级。成对力具有高度各向异性,具有拉长的涨落椭球体。残留力-残留力可以是吸引力或排斥力(尽管更有可能具有吸引力),形成某种由复杂的力网络稳定的张拉整体结构。能够理解和预测相互作用网络为合理的药物设计和揭示分子识别机制提供了基础。































 京公网安备 11010802027423号
京公网安备 11010802027423号