当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
CHARMM36 All-Atom Gas Model for Lipid Nanobubble Simulation
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-09-11 , DOI: 10.1021/acs.jcim.4c01027 Xiu Li 1 , Yuan He 1 , Yuxuan Wang 1 , Kaidong Lin 1 , Xubo Lin 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-09-11 , DOI: 10.1021/acs.jcim.4c01027 Xiu Li 1 , Yuan He 1 , Yuxuan Wang 1 , Kaidong Lin 1 , Xubo Lin 1
Affiliation
|
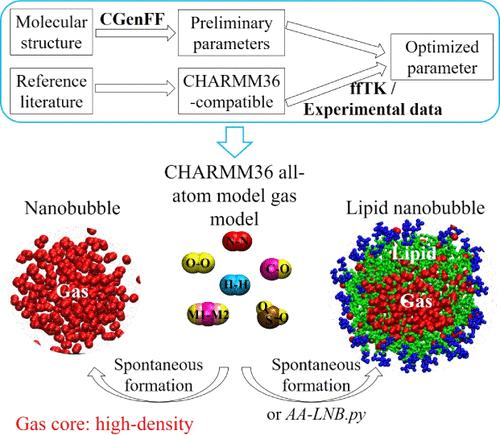
Lipid nanobubbles with different gas cores may integrate the biocompatibility of lipids, powerful physicochemical properties of nanobubbles, and therapeutic effects of gas molecules, which thus promote enormous biomedical applications such as ultrasound molecular imaging, gene/drug delivery, and gas therapy. In order for further more precise applications, the exact molecular mechanisms for the interactions between lipid nanobubbles and biological systems should be studied. Molecular dynamics (MD) simulation provides a powerful computational tool for this purpose. However, previous state-of-the-art MD simulations of free gas nanobubble/lipid nanobubble employed the vacuum as their gas cores, which is not suitable for studying the interactions between functional lipid nanobubbles and biological systems and revealing the biological roles of gas molecules. Hence, in this work, we developed and optimized the CHARMM36 all-atom gas parameters for six gases including N2, O2, H2, CO, CO2, and SO2, which accurately reproduced the gas density at different pressures as well as the spontaneous formation of gas nanobubbles. Subsequent applications of these gas parameters for lipid nanobubble simulations also reproduced the self-assembly process of the lipid nanobubble. We further developed a Python script to generate all-atom lipid nanobubble simulation systems, which was proven to be efficient for all-atom MD simulations of lipid nanobubbles and to be able to capture the exact dynamics of gas molecules at the gas–lipid and lipid–water interfaces of the lipid nanobubble. In summary, the all-atom gas models proposed in this work are suitable for simulating free gas nanobubbles and lipid nanobubbles, which are supposed to overcome the shortcomings of previous state-of-the-art MD simulations with the vacuum replacing the gas core and play key roles in revealing the molecular-level interactions between lipid nanobubbles and biological systems.
中文翻译:

CHARMM36 用于脂质纳米气泡模拟的全原子气体模型
具有不同气核的脂质纳米气泡可以整合脂质的生物相容性、纳米气泡强大的物理化学特性和气体分子的治疗作用,从而促进巨大的生物医学应用,如超声分子成像、基因/药物递送和气体疗法。为了实现更精确的应用,应研究脂质纳米气泡与生物系统之间相互作用的确切分子机制。分子动力学 (MD) 仿真为此提供了强大的计算工具。然而,以前最先进的自由气体纳米气泡/脂质纳米气泡的 MD 模拟采用真空作为其气芯,这不适合研究功能性脂质纳米气泡与生物系统之间的相互作用和揭示气体分子的生物学作用。因此,在这项工作中,我们开发和优化了 N2、O2、H2、CO、CO2 和 SO2 等六种气体的CHARMM36全原子气体参数,准确再现了不同压力下的气体密度以及气体纳米气泡的自发形成。这些气体参数随后在脂质纳米气泡模拟中的应用也再现了脂质纳米气泡的自组装过程。我们进一步开发了一个 Python 脚本来生成全原子脂质纳米气泡模拟系统,该系统被证明对于脂质纳米气泡的全原子 MD 模拟是有效的,并且能够捕获脂质纳米气泡的气体-脂质和脂质-水界面处气体分子的精确动力学。 综上所述,本研究提出的全原子气体模型适用于模拟自由气体纳米气泡和脂质纳米气泡,旨在克服以前最先进的 MD 模拟的缺点,真空取代了气芯,并在揭示脂质纳米气泡与生物系统之间的分子水平相互作用方面发挥着关键作用。
更新日期:2024-09-11
中文翻译:

CHARMM36 用于脂质纳米气泡模拟的全原子气体模型
具有不同气核的脂质纳米气泡可以整合脂质的生物相容性、纳米气泡强大的物理化学特性和气体分子的治疗作用,从而促进巨大的生物医学应用,如超声分子成像、基因/药物递送和气体疗法。为了实现更精确的应用,应研究脂质纳米气泡与生物系统之间相互作用的确切分子机制。分子动力学 (MD) 仿真为此提供了强大的计算工具。然而,以前最先进的自由气体纳米气泡/脂质纳米气泡的 MD 模拟采用真空作为其气芯,这不适合研究功能性脂质纳米气泡与生物系统之间的相互作用和揭示气体分子的生物学作用。因此,在这项工作中,我们开发和优化了 N2、O2、H2、CO、CO2 和 SO2 等六种气体的CHARMM36全原子气体参数,准确再现了不同压力下的气体密度以及气体纳米气泡的自发形成。这些气体参数随后在脂质纳米气泡模拟中的应用也再现了脂质纳米气泡的自组装过程。我们进一步开发了一个 Python 脚本来生成全原子脂质纳米气泡模拟系统,该系统被证明对于脂质纳米气泡的全原子 MD 模拟是有效的,并且能够捕获脂质纳米气泡的气体-脂质和脂质-水界面处气体分子的精确动力学。 综上所述,本研究提出的全原子气体模型适用于模拟自由气体纳米气泡和脂质纳米气泡,旨在克服以前最先进的 MD 模拟的缺点,真空取代了气芯,并在揭示脂质纳米气泡与生物系统之间的分子水平相互作用方面发挥着关键作用。































 京公网安备 11010802027423号
京公网安备 11010802027423号