Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Directly Characterizing the Capture Radius of Tethered Double-Stranded DNA by Single-Molecule Nanopipette Manipulation
ACS Nano ( IF 15.8 ) Pub Date : 2024-09-12 , DOI: 10.1021/acsnano.4c05605 Bohua Yin 1, 2 , Shaoxi Fang 2 , Bin Wu 3 , Wenhao Ma 2 , Daming Zhou 2 , Yajie Yin 2 , Rong Tian 2 , Shixuan He 2 , Jian-An Huang 4 , Wanyi Xie 2 , Xing-Hua Zhang 3 , Zuobin Wang 1 , Deqiang Wang 1, 2
ACS Nano ( IF 15.8 ) Pub Date : 2024-09-12 , DOI: 10.1021/acsnano.4c05605 Bohua Yin 1, 2 , Shaoxi Fang 2 , Bin Wu 3 , Wenhao Ma 2 , Daming Zhou 2 , Yajie Yin 2 , Rong Tian 2 , Shixuan He 2 , Jian-An Huang 4 , Wanyi Xie 2 , Xing-Hua Zhang 3 , Zuobin Wang 1 , Deqiang Wang 1, 2
Affiliation
|
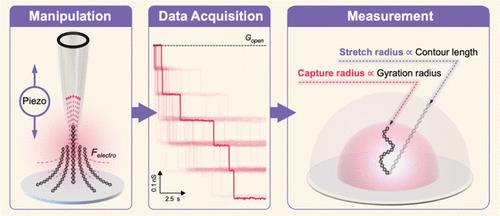
The tethered molecule exhibits characteristics of both free and fixed states, with the electrodynamics involved in its diffusion, electrophoresis, and stretching processes still not fully understood. We developed a Single-Molecule Manipulation, Identification, and Length Examination (SMILE) system by integrating piezoelectric devices with nanopipettes. This system enabled successful capture and stretching of tethered double-stranded DNA within the nanopore. Our research unveiled distinct capture (rcapture) and stretch radii (rstretch) surrounding the DNA’s anchor point. Notably, consistent ratios of capture radius for DNA of varying lengths (2k, 4k, and 6k base pairs) were observed across different capturing voltages, approximately 1:1.4:1.83, showing a resemblance to their gyration radius ratios. However, the ratios of stretch radius are consistent to their contour length (L0), with the stretching ratio (rstretch/L0) increasing from 70 to 90% as the voltage rose from 100 to 1000 mV. Additionally, through numerical simulations, we identified the origin of capture and stretch radii, determined by the entropic elasticity-induced capture barrier and the electric field-dominant escape barrier. This research introduces an innovative methodology and outlines research perspectives for a comprehensive exploration of the conformational, electrical, and diffusion characteristics of tethered molecules.
中文翻译:

通过单分子纳米移液器操作直接表征栓系双链 DNA 的捕获半径
束缚分子表现出自由态和固定态的特性,其扩散、电泳和拉伸过程所涉及的电动力学仍未完全了解。我们通过将压电器件与纳米移液器集成,开发了一种单分子操作、识别和长度检查 (SMILE) 系统。该系统能够在纳米孔内成功捕获和拉伸栓系双链 DNA。我们的研究揭示了围绕 DNA 锚点的不同捕获 (rcapture) 和拉伸半径 (rstretch)。值得注意的是,在不同的捕获电压下观察到不同长度(2k、4k 和 6k 碱基对)的 DNA 捕获半径比率一致,约为 1:1.4:1.83,显示出与它们的回转半径比率相似。然而,拉伸半径的比率与它们的轮廓长度 (L0) 一致,当电压从 100 mV 上升到 1000 mV 时,拉伸比率 (rstretch/L0) 从 70% 增加到 90%。此外,通过数值模拟,我们确定了捕获和拉伸半径的来源,由熵弹性诱导捕获屏障和电场主导的逃逸屏障决定。本研究介绍了一种创新方法,并概述了全面探索栓系分子的构象、电学和扩散特性的研究前景。
更新日期:2024-09-12
中文翻译:

通过单分子纳米移液器操作直接表征栓系双链 DNA 的捕获半径
束缚分子表现出自由态和固定态的特性,其扩散、电泳和拉伸过程所涉及的电动力学仍未完全了解。我们通过将压电器件与纳米移液器集成,开发了一种单分子操作、识别和长度检查 (SMILE) 系统。该系统能够在纳米孔内成功捕获和拉伸栓系双链 DNA。我们的研究揭示了围绕 DNA 锚点的不同捕获 (rcapture) 和拉伸半径 (rstretch)。值得注意的是,在不同的捕获电压下观察到不同长度(2k、4k 和 6k 碱基对)的 DNA 捕获半径比率一致,约为 1:1.4:1.83,显示出与它们的回转半径比率相似。然而,拉伸半径的比率与它们的轮廓长度 (L0) 一致,当电压从 100 mV 上升到 1000 mV 时,拉伸比率 (rstretch/L0) 从 70% 增加到 90%。此外,通过数值模拟,我们确定了捕获和拉伸半径的来源,由熵弹性诱导捕获屏障和电场主导的逃逸屏障决定。本研究介绍了一种创新方法,并概述了全面探索栓系分子的构象、电学和扩散特性的研究前景。































 京公网安备 11010802027423号
京公网安备 11010802027423号