Diabetologia ( IF 8.4 ) Pub Date : 2024-09-06 , DOI: 10.1007/s00125-024-06261-x Nicholas A Wachowski 1, 2 , James A Pippin 1, 2 , Keith Boehm 1, 2 , Sumei Lu 1, 2 , Michelle E Leonard 1, 2 , Elisabetta Manduchi 1, 2, 3 , Ursula W Parlin 1, 2, 4 , Martin Wabitsch 5 , Alessandra Chesi 6 , Andrew D Wells 1, 6, 7, 8 , Struan F A Grant 1, 2, 3, 9, 10 , Matthew C Pahl 1, 2
|
Aims/hypothesis
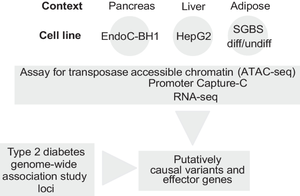
Genome-wide association studies (GWAS) have identified hundreds of type 2 diabetes loci, with the vast majority of signals located in non-coding regions; as a consequence, it remains largely unclear which ‘effector’ genes these variants influence. Determining these effector genes has been hampered by the relatively challenging cellular settings in which they are hypothesised to confer their effects.
Methods
To implicate such effector genes, we elected to generate and integrate high-resolution promoter-focused Capture-C, assay for transposase-accessible chromatin with sequencing (ATAC-seq) and RNA-seq datasets to characterise chromatin and expression profiles in multiple cell lines relevant to type 2 diabetes for subsequent functional follow-up analyses: EndoC-BH1 (pancreatic beta cell), HepG2 (hepatocyte) and Simpson–Golabi–Behmel syndrome (SGBS; adipocyte).
Results
The subsequent variant-to-gene analysis implicated 810 candidate effector genes at 370 type 2 diabetes risk loci. Using partitioned linkage disequilibrium score regression, we observed enrichment for type 2 diabetes and fasting glucose GWAS loci in promoter-connected putative cis-regulatory elements in EndoC-BH1 cells as well as fasting insulin GWAS loci in SGBS cells. Moreover, as a proof of principle, when we knocked down expression of the SMCO4 gene in EndoC-BH1 cells, we observed a statistically significant increase in insulin secretion.
Conclusions/interpretation
These results provide a resource for comparing tissue-specific data in tractable cellular models as opposed to relatively challenging primary cell settings.
Data availability
Raw and processed next-generation sequencing data for EndoC-BH1, HepG2, SGBS_undiff and SGBS_diff cells are deposited in GEO under the Superseries accession GSE262484. Promoter-focused Capture-C data are deposited under accession GSE262496. Hi-C data are deposited under accession GSE262481. Bulk ATAC-seq data are deposited under accession GSE262479. Bulk RNA-seq data are deposited under accession GSE262480.
Graphical Abstract
中文翻译:

使用以启动子为中心的 Capture-C 将 2 型糖尿病效应基因与相关代谢细胞模型联系起来
目标/假设
全基因组关联研究 (GWAS) 已经确定了数百个 2 型糖尿病位点,其中绝大多数信号位于非编码区;因此,目前尚不清楚这些变体影响了哪些 “效应 ”基因。确定这些效应基因受到相对具有挑战性的细胞环境的阻碍,在这些环境中,它们被假设以赋予它们的作用。
方法
为了涉及此类效应基因,我们选择生成并整合高分辨率以启动子为中心的 Capture-C、转座酶可及染色质检测与测序 (ATAC-seq) 和 RNA-seq 数据集,以表征与 2 型糖尿病相关的多个细胞系中的染色质和表达谱,用于后续功能随访分析:EndoC-BH1(胰腺 β 细胞)、HepG2(肝细胞)和 Simpson-Golabi-Behmel 综合征(SGBS;脂肪细胞)。
结果
随后的变异到基因分析表明,810 个候选效应基因位于 370 个 2 型糖尿病风险位点。使用分区连锁不平衡评分回归,我们观察到 EndoC-BH1 细胞中启动子连接的推定顺式调节元件中 2 型糖尿病和空腹血糖 GWAS 基因座以及 SGBS 细胞中空腹胰岛素 GWAS 基因座的富集。此外,作为原理证明,当我们敲低 EndoC-BH1 细胞中 SMCO4 基因的表达时,我们观察到胰岛素分泌的统计学显着增加。
结论/解释
这些结果为比较易处理细胞模型中的组织特异性数据提供了资源,而不是相对具有挑战性的原代细胞设置。
数据可用性
EndoC-BH1、HepG2、SGBS_undiff 和 SGBS_diff 细胞的原始和处理的下一代测序数据在超系列登录GSE262484下沉积在 GEO 中。以启动子为中心的 Capture-C 数据以 Accession GSE262496 保存。Hi-C 数据以 Accession GSE262481 的形式存储。批量 ATAC-seq 数据以 GSE262479 方式存放。大量 RNA-seq 数据以 Accession GSE262480 保存。































 京公网安备 11010802027423号
京公网安备 11010802027423号