当前位置:
X-MOL 学术
›
Mol. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
In Vivo Cross-Linking Sheds Light on the Salmonella Divisome in Which PBP3 and PBP3SAL Compete for Occupancy
Molecular Microbiology ( IF 2.6 ) Pub Date : 2024-09-04 , DOI: 10.1111/mmi.15309 Sónia Castanheira 1 , David López-Escarpa 1 , Alberto Paradela 2 , Francisco García-Del Portillo 1
Molecular Microbiology ( IF 2.6 ) Pub Date : 2024-09-04 , DOI: 10.1111/mmi.15309 Sónia Castanheira 1 , David López-Escarpa 1 , Alberto Paradela 2 , Francisco García-Del Portillo 1
Affiliation
|
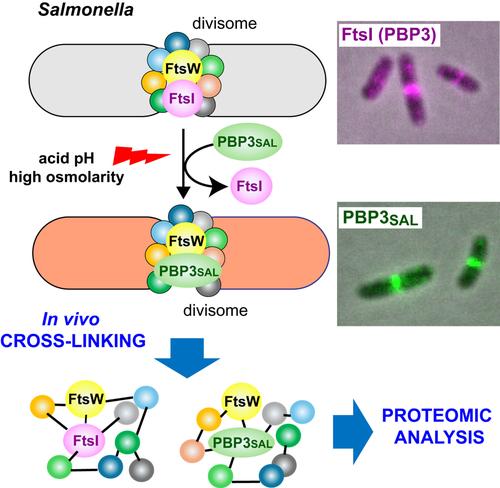
Bacterial cell division is orchestrated by proteins that assemble in dynamic complexes collectively known as the divisome. Essential monofunctional enzymes with glycosyltransferase or transpeptidase (TPase) activities, FtsW and FtsI respectively, engage in the synthesis of septal peptidoglycan (sPG). Enigmatically, Salmonella has two TPases that can promote cell division independently: FtsI (PBP3) and the pathogen-specific paralogue PBP3SAL. How Salmonella regulates the assembly of the sPG synthase complex with these two TPases, is unknown. Here, we characterized Salmonella division complexes in wild-type cells and isogenic mutants lacking PBP3 or PBP3SAL. The complexes were cross-linked in vivo and pulled down with antibodies recognizing each enzyme. Proteomics of the immunoprecipitates showed that PBP3 and PBP3SAL do not extensively cross-link in wild type cells, supporting the presence of independent complexes. More than 40 proteins cross-link in complexes in which these two TPases are present. Those identified with high scores include FtsA, FtsK, FtsQLB, FtsW, PBP1B, SPOR domain-containing proteins (FtsN, DedD, RlpA, DamX), amidase activators (FtsX, EnvC, NlpD) and Tol-Pal proteins. Other cross-linked proteins are the protease Prc, the elongasome TPase PBP2 and, D,D-endo- and D,D-carboxypeptidases. PBP3 and PBP3SAL localize at midcell and compete for occupying the division complex in response to environmental cues. Thus, a catalytic-dead PBP3SAL-S300A variant impairs cell division in a high osmolarity and acidic condition in which it is produced at levels exceeding those of PBP3. Salmonella may therefore exploit an ‘adjustable’ divisome to exchange TPases for ensuring cell division in distinct environments and, in this manner, expand its colonization capacities.
中文翻译:

体内交联揭示了 PBP3 和 PBP3SAL 争夺占有率的沙门氏菌分裂体
细菌细胞分裂是由蛋白质精心编排的,这些蛋白质组装成统称为分裂体的动态复合物。具有糖基转移酶或转肽酶 (TPase) 活性的必需单功能酶 FtsW 和 FtsI 分别参与隔膜肽聚糖 (sPG) 的合成。神秘地,沙门氏菌有两种可以独立促进细胞分裂的 TPase:FtsI (PBP3) 和病原体特异性旁系同源物 PBP3SAL。沙门氏菌如何调节 sPG 合酶复合物与这两种 TPase 的组装尚不清楚。在这里,我们表征了野生型细胞和缺乏 PBP3 或 PBP3SAL 的同基因突变体中的沙门氏菌分裂复合物。复合物在体内交联,并用识别每种酶的抗体拉下。免疫沉淀物的蛋白质组学表明,PBP3 和 PBP3SAL 在野生型细胞中不广泛交联,支持独立复合物的存在。超过 40 种蛋白质在存在这两种 TPase 的复合物中交联。高分鉴定的包括 FtsA、FtsK、FtsQLB、FtsW、PBP1B、SPOR 结构域包含的蛋白质 (FtsN、DedD、RlpA、DamX)、酰胺酶激活剂 (FtsX、EnvC、NlpD) 和 Tol-Pal 蛋白。其他交联蛋白是蛋白酶 Prc、延长体 TPase PBP2 以及 D,D-内切酶和 D,D-羧肽酶。PBP3 和 PBP3SAL 定位于中细胞,并响应环境线索竞争占据分裂复合物。因此,催化死亡的 PBP3 SAL-S300A 变体在高渗透压和酸性条件下会损害细胞分裂,其中产生的水平超过 PBP3。 因此,沙门氏菌可以利用“可调”的二分体来交换 TPase,以确保在不同环境中进行细胞分裂,并以这种方式扩大其定植能力。
更新日期:2024-09-04
中文翻译:

体内交联揭示了 PBP3 和 PBP3SAL 争夺占有率的沙门氏菌分裂体
细菌细胞分裂是由蛋白质精心编排的,这些蛋白质组装成统称为分裂体的动态复合物。具有糖基转移酶或转肽酶 (TPase) 活性的必需单功能酶 FtsW 和 FtsI 分别参与隔膜肽聚糖 (sPG) 的合成。神秘地,沙门氏菌有两种可以独立促进细胞分裂的 TPase:FtsI (PBP3) 和病原体特异性旁系同源物 PBP3SAL。沙门氏菌如何调节 sPG 合酶复合物与这两种 TPase 的组装尚不清楚。在这里,我们表征了野生型细胞和缺乏 PBP3 或 PBP3SAL 的同基因突变体中的沙门氏菌分裂复合物。复合物在体内交联,并用识别每种酶的抗体拉下。免疫沉淀物的蛋白质组学表明,PBP3 和 PBP3SAL 在野生型细胞中不广泛交联,支持独立复合物的存在。超过 40 种蛋白质在存在这两种 TPase 的复合物中交联。高分鉴定的包括 FtsA、FtsK、FtsQLB、FtsW、PBP1B、SPOR 结构域包含的蛋白质 (FtsN、DedD、RlpA、DamX)、酰胺酶激活剂 (FtsX、EnvC、NlpD) 和 Tol-Pal 蛋白。其他交联蛋白是蛋白酶 Prc、延长体 TPase PBP2 以及 D,D-内切酶和 D,D-羧肽酶。PBP3 和 PBP3SAL 定位于中细胞,并响应环境线索竞争占据分裂复合物。因此,催化死亡的 PBP3 SAL-S300A 变体在高渗透压和酸性条件下会损害细胞分裂,其中产生的水平超过 PBP3。 因此,沙门氏菌可以利用“可调”的二分体来交换 TPase,以确保在不同环境中进行细胞分裂,并以这种方式扩大其定植能力。






























 京公网安备 11010802027423号
京公网安备 11010802027423号