当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Comparative genomics identifies key adaptive traits of sponge‐associated microbial symbionts
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-09-04 , DOI: 10.1111/1462-2920.16690 Paul A O'Brien 1, 2, 3, 4 , Steven J Robbins 4 , Shangjin Tan 5, 6 , Laura Rix 4 , David J Miller 7, 8 , Nicole S Webster 2, 4, 9 , Guojie Zhang 10, 11 , David G Bourne 1, 2, 3
Environmental Microbiology ( IF 4.3 ) Pub Date : 2024-09-04 , DOI: 10.1111/1462-2920.16690 Paul A O'Brien 1, 2, 3, 4 , Steven J Robbins 4 , Shangjin Tan 5, 6 , Laura Rix 4 , David J Miller 7, 8 , Nicole S Webster 2, 4, 9 , Guojie Zhang 10, 11 , David G Bourne 1, 2, 3
Affiliation
|
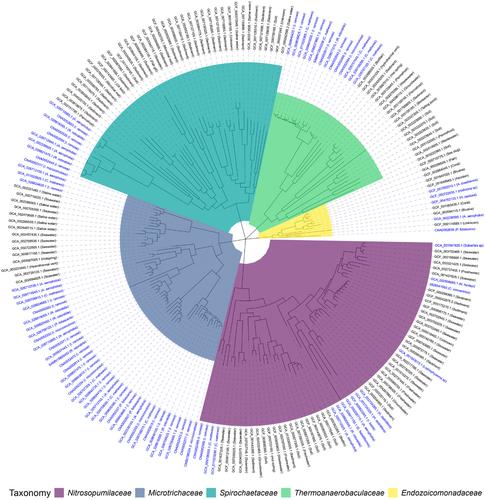
Sponge microbiomes are often highly diverse making it difficult to determine which lineages are important for maintaining host health and homeostasis. Characterising genomic traits associated with symbiosis can improve our knowledge of which lineages have adapted to their host and what functions they might provide. Here we examined five microbial families associated with sponges that have previously shown evidence of cophylogeny, including Endozoicomonadaceae, Nitrosopumilaceae, Spirochaetaceae, Microtrichaceae and Thermoanaerobaculaceae , to better understand the mechanisms behind their symbiosis. We compared sponge‐associated genomes to genomes found in other environments and found that sponge‐specific clades were enriched in genes encoding many known mechanisms for symbiont survival, such as avoiding phagocytosis and defence against foreign genetic elements. We expand on previous knowledge to show that glycosyl hydrolases with sulfatases and sulfotransferases likely form multienzyme degradation pathways to break and remodel sulfated polysaccharides and reveal an enrichment in superoxide dismutase that may prevent damage from free oxygen radicals produced by the host. Finally, we identified novel traits in sponge‐associated symbionts, such as urea metabolism in Spirochaetaceae which was previously shown to be rare in the phylum Spirochaetota. These results identify putative mechanisms by which symbionts have adapted to living in association with sponges.
中文翻译:

比较基因组学识别海绵相关微生物共生体的关键适应性特征
海绵微生物组通常高度多样化,因此很难确定哪些谱系对于维持宿主健康和体内平衡很重要。表征与共生相关的基因组特征可以提高我们对哪些谱系已经适应其宿主以及它们可能提供哪些功能的了解。在这里,我们检查了与海绵相关的五个微生物科,这些微生物科先前已显示出共系统发育的证据,包括Endozoicomonadaceae、Nitrosopumilaceae、Spirochaetaceae、Microtrichaceae和Thermoanaerobaculaceae,以更好地了解它们共生背后的机制。我们将海绵相关基因组与其他环境中发现的基因组进行比较,发现海绵特异性进化枝富含编码许多已知共生体生存机制的基因,例如避免吞噬作用和防御外来遗传元件。我们扩展了先前的知识,表明糖基水解酶与硫酸酯酶和磺基转移酶可能形成多酶降解途径来破坏和重塑硫酸化多糖,并揭示超氧化物歧化酶的富集,可以防止宿主产生的自由基造成的损害。最后,我们发现了与海绵相关的共生体的新特征,例如螺旋体科的尿素代谢,这在之前的研究中被证明在螺旋体门中是罕见的。这些结果确定了共生体适应与海绵共存的假定机制。
更新日期:2024-09-04
中文翻译:

比较基因组学识别海绵相关微生物共生体的关键适应性特征
海绵微生物组通常高度多样化,因此很难确定哪些谱系对于维持宿主健康和体内平衡很重要。表征与共生相关的基因组特征可以提高我们对哪些谱系已经适应其宿主以及它们可能提供哪些功能的了解。在这里,我们检查了与海绵相关的五个微生物科,这些微生物科先前已显示出共系统发育的证据,包括Endozoicomonadaceae、Nitrosopumilaceae、Spirochaetaceae、Microtrichaceae和Thermoanaerobaculaceae,以更好地了解它们共生背后的机制。我们将海绵相关基因组与其他环境中发现的基因组进行比较,发现海绵特异性进化枝富含编码许多已知共生体生存机制的基因,例如避免吞噬作用和防御外来遗传元件。我们扩展了先前的知识,表明糖基水解酶与硫酸酯酶和磺基转移酶可能形成多酶降解途径来破坏和重塑硫酸化多糖,并揭示超氧化物歧化酶的富集,可以防止宿主产生的自由基造成的损害。最后,我们发现了与海绵相关的共生体的新特征,例如螺旋体科的尿素代谢,这在之前的研究中被证明在螺旋体门中是罕见的。这些结果确定了共生体适应与海绵共存的假定机制。




















































 京公网安备 11010802027423号
京公网安备 11010802027423号