当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Predicting Small Molecule Binding Nucleotides in RNA Structures Using RNA Surface Topography
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-09-04 , DOI: 10.1021/acs.jcim.4c01264 Jiaming Gao 1 , Haoquan Liu 1 , Chen Zhuo 1 , Chengwei Zeng 1 , Yunjie Zhao 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-09-04 , DOI: 10.1021/acs.jcim.4c01264 Jiaming Gao 1 , Haoquan Liu 1 , Chen Zhuo 1 , Chengwei Zeng 1 , Yunjie Zhao 1
Affiliation
|
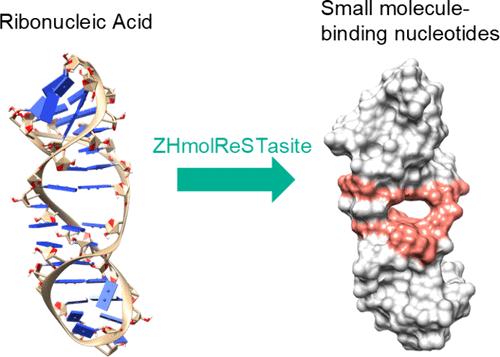
RNA small molecule interactions play a crucial role in drug discovery and inhibitor design. Identifying RNA small molecule binding nucleotides is essential and requires methods that exhibit a high predictive ability to facilitate drug discovery and inhibitor design. Existing methods can predict the binding nucleotides of simple RNA structures, but it is hard to predict binding nucleotides in complex RNA structures with junctions. To address this limitation, we developed a new deep learning model based on spatial correlation, ZHmolReSTasite, which can accurately predict binding nucleotides of small and large RNA with junctions. We utilize RNA surface topography to consider the spatial correlation, characterizing nucleotides from sequence and tertiary structures to learn a high-level representation. Our method outperforms existing methods for benchmark test sets composed of simple RNA structures, achieving precision values of 72.9% on TE18 and 76.7% on RB9 test sets. For a challenging test set composed of RNA structures with junctions, our method outperforms the second best method by 11.6% in precision. Moreover, ZHmolReSTasite demonstrates robustness regarding the predicted RNA structures. In summary, ZHmolReSTasite successfully incorporates spatial correlation, outperforms previous methods on small and large RNA structures using RNA surface topography, and can provide valuable insights into RNA small molecule prediction and accelerate RNA inhibitor design.
中文翻译:

使用 RNA 表面形貌预测 RNA 结构中的小分子结合核苷酸
RNA 小分子相互作用在药物发现和抑制剂设计中发挥着至关重要的作用。识别 RNA 小分子结合核苷酸至关重要,并且需要具有高预测能力的方法来促进药物发现和抑制剂设计。现有方法可以预测简单RNA结构的结合核苷酸,但很难预测具有连接的复杂RNA结构中的结合核苷酸。为了解决这个限制,我们开发了一种基于空间相关性的新深度学习模型ZHmolReSTasite,它可以准确预测小RNA和大RNA与连接点的结合核苷酸。我们利用 RNA 表面形貌来考虑空间相关性,从序列和三级结构中表征核苷酸以学习高级表示。我们的方法优于由简单 RNA 结构组成的基准测试集的现有方法,在 TE18 上实现了 72.9% 的精度值,在 RB9 测试集上实现了 76.7% 的精度值。对于由带有连接的 RNA 结构组成的具有挑战性的测试集,我们的方法在精度上比第二好的方法高出 11.6%。此外,ZHmolReSTasite 展示了预测 RNA 结构的稳健性。总之,ZHmolReSTasite 成功地整合了空间相关性,优于之前使用 RNA 表面形貌处理小和大 RNA 结构的方法,并且可以为 RNA 小分子预测提供有价值的见解并加速 RNA 抑制剂设计。
更新日期:2024-09-04
中文翻译:

使用 RNA 表面形貌预测 RNA 结构中的小分子结合核苷酸
RNA 小分子相互作用在药物发现和抑制剂设计中发挥着至关重要的作用。识别 RNA 小分子结合核苷酸至关重要,并且需要具有高预测能力的方法来促进药物发现和抑制剂设计。现有方法可以预测简单RNA结构的结合核苷酸,但很难预测具有连接的复杂RNA结构中的结合核苷酸。为了解决这个限制,我们开发了一种基于空间相关性的新深度学习模型ZHmolReSTasite,它可以准确预测小RNA和大RNA与连接点的结合核苷酸。我们利用 RNA 表面形貌来考虑空间相关性,从序列和三级结构中表征核苷酸以学习高级表示。我们的方法优于由简单 RNA 结构组成的基准测试集的现有方法,在 TE18 上实现了 72.9% 的精度值,在 RB9 测试集上实现了 76.7% 的精度值。对于由带有连接的 RNA 结构组成的具有挑战性的测试集,我们的方法在精度上比第二好的方法高出 11.6%。此外,ZHmolReSTasite 展示了预测 RNA 结构的稳健性。总之,ZHmolReSTasite 成功地整合了空间相关性,优于之前使用 RNA 表面形貌处理小和大 RNA 结构的方法,并且可以为 RNA 小分子预测提供有价值的见解并加速 RNA 抑制剂设计。

































 京公网安备 11010802027423号
京公网安备 11010802027423号