当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Enhancing protein-ligand binding affinity prediction through sequential fusion of graph and convolutional neural networks
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-09-02 , DOI: 10.1002/jcc.27499 Yimin Yang 1 , Ruiqin Zhang 2 , Zijing Lin 1, 3
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2024-09-02 , DOI: 10.1002/jcc.27499 Yimin Yang 1 , Ruiqin Zhang 2 , Zijing Lin 1, 3
Affiliation
|
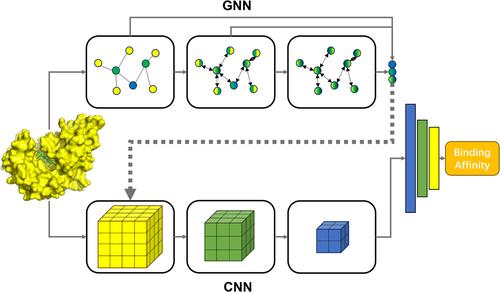
Predicting protein-ligand binding affinity is a crucial and challenging task in structure-based drug discovery. With the accumulation of complex structures and binding affinity data, various machine-learning scoring functions, particularly those based on deep learning, have been developed for this task, exhibiting superiority over their traditional counterparts. A fusion model sequentially connecting a graph neural network (GNN) and a convolutional neural network (CNN) to predict protein-ligand binding affinity is proposed in this work. In this model, the intermediate outputs of the GNN layers, as supplementary descriptors of atomic chemical environments at different levels, are concatenated with the input features of CNN. The model demonstrates a noticeable improvement in performance on CASF-2016 benchmark compared to its constituent CNN models. The generalization ability of the model is evaluated by setting a series of thresholds for ligand extended-connectivity fingerprint similarity or protein sequence similarity between the training and test sets. Masking experiment reveals that model can capture key interaction regions. Furthermore, the fusion model is applied to a virtual screening task for a novel target, PI5P4Kα. The fusion strategy significantly improves the ability of the constituent CNN model to identify active compounds. This work offers a novel approach to enhancing the accuracy of deep learning models in predicting binding affinity through fusion strategies.
中文翻译:

通过图和卷积神经网络的顺序融合增强蛋白质-配体结合亲和力预测
预测蛋白质-配体结合亲和力是基于结构的药物发现中一项关键且具有挑战性的任务。随着复杂结构和结合亲和力数据的积累,各种机器学习评分函数,特别是基于深度学习的评分函数,已经为这项任务开发了,表现出优于传统函数的优点。这项工作提出了一种按顺序连接图神经网络 (GNN) 和卷积神经网络 (CNN) 的融合模型来预测蛋白质-配体结合亲和力。在这个模型中,GNN 层的中间输出作为不同能级原子化学环境的补充描述符,与 CNN 的输入特征连接起来。与其组成 CNN 模型相比,该模型在 CASF-2016 基准测试中的性能有了显著提高。通过为训练集和测试集之间的配体扩展连接指纹相似性或蛋白质序列相似性设置一系列阈值来评估模型的泛化能力。掩蔽实验表明,模型可以捕获关键的交互区域。此外,融合模型应用于新靶点 PI5P4Kα 的虚拟筛选任务。融合策略显著提高了组成 CNN 模型识别活性化合物的能力。这项工作提供了一种新的方法,可以提高深度学习模型通过融合策略预测结合亲和力的准确性。
更新日期:2024-09-02
中文翻译:

通过图和卷积神经网络的顺序融合增强蛋白质-配体结合亲和力预测
预测蛋白质-配体结合亲和力是基于结构的药物发现中一项关键且具有挑战性的任务。随着复杂结构和结合亲和力数据的积累,各种机器学习评分函数,特别是基于深度学习的评分函数,已经为这项任务开发了,表现出优于传统函数的优点。这项工作提出了一种按顺序连接图神经网络 (GNN) 和卷积神经网络 (CNN) 的融合模型来预测蛋白质-配体结合亲和力。在这个模型中,GNN 层的中间输出作为不同能级原子化学环境的补充描述符,与 CNN 的输入特征连接起来。与其组成 CNN 模型相比,该模型在 CASF-2016 基准测试中的性能有了显著提高。通过为训练集和测试集之间的配体扩展连接指纹相似性或蛋白质序列相似性设置一系列阈值来评估模型的泛化能力。掩蔽实验表明,模型可以捕获关键的交互区域。此外,融合模型应用于新靶点 PI5P4Kα 的虚拟筛选任务。融合策略显著提高了组成 CNN 模型识别活性化合物的能力。这项工作提供了一种新的方法,可以提高深度学习模型通过融合策略预测结合亲和力的准确性。































 京公网安备 11010802027423号
京公网安备 11010802027423号