Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
In silico selection of aptamers against SARS-CoV-2
Analyst ( IF 3.6 ) Pub Date : 2024-09-02 , DOI: 10.1039/d4an00812j Amir Muhaimin Akmal Shukri 1, 2 , Seok Mui Wang 2, 3, 4, 5 , Chaoli Feng 1 , Suet Lin Chia 6, 7, 8 , Siti Farah Alwani Mohd Nawi 3 , Marimuthu Citartan 1
Analyst ( IF 3.6 ) Pub Date : 2024-09-02 , DOI: 10.1039/d4an00812j Amir Muhaimin Akmal Shukri 1, 2 , Seok Mui Wang 2, 3, 4, 5 , Chaoli Feng 1 , Suet Lin Chia 6, 7, 8 , Siti Farah Alwani Mohd Nawi 3 , Marimuthu Citartan 1
Affiliation
|
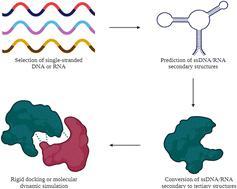
Aptamers are molecular recognition elements that have been extensively deployed in a wide array of applications ranging from diagnostics to therapeutics. Due to their unique properties as compared to antibodies, aptamers were also largely isolated during the COVID-19 pandemic for multiple purposes. Typically generated by conventional SELEX, the inherent drawbacks of the process including the time-consuming, cumbersome and resource-intensive nature catalysed the move to adopt in silico approaches to isolate aptamers. Impressive performances of these in silico-derived aptamers in their respective assays have been documented thus far, bearing testimony to the huge potential of the in silico approaches, akin to the traditional SELEX in isolating aptamers. In this study, we provide an overview of the in silico selection of aptamers against SARS-CoV-2 by providing insights into the basic steps involved, which comprise the selection of the initial single-stranded nucleic acids, determination of the secondary and tertiary structures and in silico approaches that include both rigid docking and molecular dynamics simulations. The different approaches involving aptamers against SARS-CoV-2 were illuminated and the need to verify these aptamers by experimental validation was also emphasized. Cognizant of the need to continuously improve aptamers, the strategies embraced thus far for post-in silico selection modifications were enumerated. Shedding light on the steps involved in the in silico selection can set the stage for further improvisation to augment the functionalities of the aptamers in the future.
中文翻译:

针对 SARS-CoV-2 的适体的计算机选择
适体是分子识别元件,已广泛应用于从诊断到治疗的各种应用中。由于与抗体相比具有独特的特性,适体在 COVID-19 大流行期间也基本上被分离出来用于多种目的。通常由传统的 SELEX 产生,该过程的固有缺点,包括耗时、繁琐和资源密集的性质,促使人们采用计算机方法来分离适体。迄今为止,这些计算机衍生的适体在各自的测定中的令人印象深刻的性能已被记录,证明了计算机方法的巨大潜力,类似于分离适体的传统SELEX。在本研究中,我们通过深入了解所涉及的基本步骤(包括初始单链核酸的选择、二级和三级结构的确定),概述了针对 SARS-CoV-2 的适体的计算机选择以及包括刚性对接和分子动力学模拟在内的计算机方法。阐明了涉及针对 SARS-CoV-2 适体的不同方法,并强调了通过实验验证来验证这些适体的必要性。认识到不断改进适体的必要性,列举了迄今为止采用的计算机后选择修饰策略。阐明计算机选择中涉及的步骤可以为进一步的即兴创作奠定基础,以增强未来适体的功能。
更新日期:2024-09-02
中文翻译:

针对 SARS-CoV-2 的适体的计算机选择
适体是分子识别元件,已广泛应用于从诊断到治疗的各种应用中。由于与抗体相比具有独特的特性,适体在 COVID-19 大流行期间也基本上被分离出来用于多种目的。通常由传统的 SELEX 产生,该过程的固有缺点,包括耗时、繁琐和资源密集的性质,促使人们采用计算机方法来分离适体。迄今为止,这些计算机衍生的适体在各自的测定中的令人印象深刻的性能已被记录,证明了计算机方法的巨大潜力,类似于分离适体的传统SELEX。在本研究中,我们通过深入了解所涉及的基本步骤(包括初始单链核酸的选择、二级和三级结构的确定),概述了针对 SARS-CoV-2 的适体的计算机选择以及包括刚性对接和分子动力学模拟在内的计算机方法。阐明了涉及针对 SARS-CoV-2 适体的不同方法,并强调了通过实验验证来验证这些适体的必要性。认识到不断改进适体的必要性,列举了迄今为止采用的计算机后选择修饰策略。阐明计算机选择中涉及的步骤可以为进一步的即兴创作奠定基础,以增强未来适体的功能。































 京公网安备 11010802027423号
京公网安备 11010802027423号