Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Full-length circRNA sequencing method using low-input RNAs and profiling of circRNAs in MPTP-PD mice on a nanopore platform
Analyst ( IF 3.6 ) Pub Date : 2024-08-28 , DOI: 10.1039/d4an00715h Ying Wang 1 , Xiaohan Li 1, 2 , Wenxiang Lu 1 , Fuyu Li 1 , Lingsong Yao 1 , Zhiyu Liu 1 , Huajuan Shi 1 , Weizhong Zhang 3 , Yunfei Bai 1
Analyst ( IF 3.6 ) Pub Date : 2024-08-28 , DOI: 10.1039/d4an00715h Ying Wang 1 , Xiaohan Li 1, 2 , Wenxiang Lu 1 , Fuyu Li 1 , Lingsong Yao 1 , Zhiyu Liu 1 , Huajuan Shi 1 , Weizhong Zhang 3 , Yunfei Bai 1
Affiliation
|
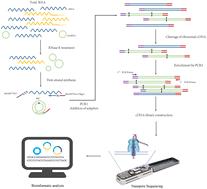
Considering the importance of accurate information of full-length (FL) transcripts in functional analysis, researchers prefer to develop new sequencing methods based on third-generation sequencing (TGS) rather than short-read sequencing. Several FL circRNA sequencing strategies have been developed. However, the current methods are inapplicable to low-biomass samples, since a large amount of total RNAs are acquired for circRNA enrichment before library preparation. In this work, we developed an effective method to detect FL circRNAs from a nanogram level (1–100 ng) of total RNAs based on a nanopore platform. Additionally, prior to the library preparation process, we added a series of 24 nt barcodes for each sample to reduce the cost and operating time. Using this method, we profiled circRNA expression in the striatum, hippocampus and cerebral cortex of a Parkinson's disease (PD) mouse model. Over 6% of reads were effective for FL circRNA identification in most datasets. Notably, a reduction in the RNA initial input resulted in a lower correlation between replicates and the detection efficiency for longer circRNA, but the lowest input (1 ng) was able to detect numerous FL circRNAs. Next, we systematically identified over 263 934 circRNAs in PD and healthy mice using the lower-input FL sequencing method, some of which came from 50.52% of PD-associated genes. Moreover, significant changes were observed in the circRNA expression pattern at an isoform level, and high-confidence protein translation evidence was predicted. Overall, we developed an effective method to characterize FL circRNAs from low-input samples and provide a comprehensive insight into the biological function of circRNAs in PD at an isoform level.
中文翻译:

使用低起始 RNA 的全长 circRNA 测序方法和在纳米孔平台上分析 MPTP-PD 小鼠中的 circRNA 分析
考虑到全长 (FL) 转录本的准确信息在功能分析中的重要性,研究人员更愿意开发基于第三代测序 (TGS) 的新测序方法,而不是短读长测序。已经开发了几种 FL circRNA 测序策略。然而,目前的方法不适用于低生物量样品,因为在文库制备之前会获得大量总 RNA 用于 circRNA 富集。在这项工作中,我们开发了一种基于纳米孔平台从纳克级 (1-100 ng) 总 RNA 中检测 FL circRNAs 的有效方法。此外,在文库制备过程之前,我们为每个样品添加了一系列 24 nt 条形码,以降低成本和操作时间。使用这种方法,我们分析了帕金森病 (PD) 小鼠模型的纹状体、海马体和大脑皮层中的 circRNA 表达。在大多数数据集中,超过 6% 的读数对 FL circRNA 鉴定有效。值得注意的是,RNA 初始起始量的减少导致重复序列与较长 circRNA 的检测效率之间的相关性降低,但最低起始量 (1 ng) 能够检测大量 FL circRNA。接下来,我们使用低起始量 FL 测序方法在 PD 和健康小鼠中系统鉴定了超过 263 934 个 circRNA,其中一些来自 50.52% 的 PD 相关基因。此外,在亚型水平上观察到 circRNA 表达模式的显着变化,并预测了高置信度的蛋白质翻译证据。总体而言,我们开发了一种有效的方法来表征低起始量样品中的 FL circRNA,并在亚型水平上全面了解 circRNAs 在 PD 中的生物学功能。
更新日期:2024-08-28
中文翻译:

使用低起始 RNA 的全长 circRNA 测序方法和在纳米孔平台上分析 MPTP-PD 小鼠中的 circRNA 分析
考虑到全长 (FL) 转录本的准确信息在功能分析中的重要性,研究人员更愿意开发基于第三代测序 (TGS) 的新测序方法,而不是短读长测序。已经开发了几种 FL circRNA 测序策略。然而,目前的方法不适用于低生物量样品,因为在文库制备之前会获得大量总 RNA 用于 circRNA 富集。在这项工作中,我们开发了一种基于纳米孔平台从纳克级 (1-100 ng) 总 RNA 中检测 FL circRNAs 的有效方法。此外,在文库制备过程之前,我们为每个样品添加了一系列 24 nt 条形码,以降低成本和操作时间。使用这种方法,我们分析了帕金森病 (PD) 小鼠模型的纹状体、海马体和大脑皮层中的 circRNA 表达。在大多数数据集中,超过 6% 的读数对 FL circRNA 鉴定有效。值得注意的是,RNA 初始起始量的减少导致重复序列与较长 circRNA 的检测效率之间的相关性降低,但最低起始量 (1 ng) 能够检测大量 FL circRNA。接下来,我们使用低起始量 FL 测序方法在 PD 和健康小鼠中系统鉴定了超过 263 934 个 circRNA,其中一些来自 50.52% 的 PD 相关基因。此外,在亚型水平上观察到 circRNA 表达模式的显着变化,并预测了高置信度的蛋白质翻译证据。总体而言,我们开发了一种有效的方法来表征低起始量样品中的 FL circRNA,并在亚型水平上全面了解 circRNAs 在 PD 中的生物学功能。































 京公网安备 11010802027423号
京公网安备 11010802027423号