当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Multitask Learning on Graph Convolutional Residual Neural Networks for Screening of Multitarget Anticancer Compounds
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-28 , DOI: 10.1021/acs.jcim.4c00643 Thanh-Hoang Nguyen-Vo 1 , Trang T T Do 1 , Binh P Nguyen 2
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-28 , DOI: 10.1021/acs.jcim.4c00643 Thanh-Hoang Nguyen-Vo 1 , Trang T T Do 1 , Binh P Nguyen 2
Affiliation
|
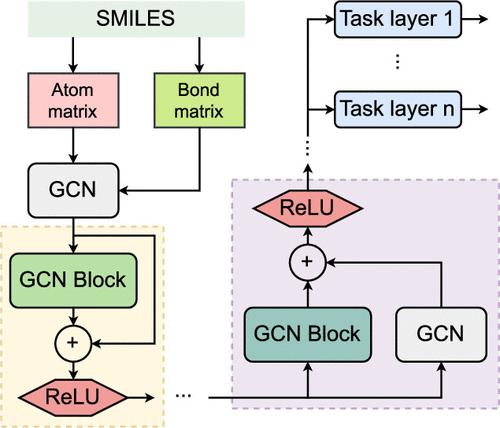
Recently, various modern experimental screening pipelines and assays have been developed to find promising anticancer drug candidates. However, it is time-consuming and almost infeasible to screen an immense number of compounds for anticancer activity via experimental approaches. To partially address this issue, several computational advances have been proposed. In this study, we present iACP-GCR, a model based on multitask learning on graph convolutional residual neural networks with two types of shortcut connections, to identify multitarget anticancer compounds. In our architecture, the graph convolutional residual neural networks are shared by all the prediction tasks before being separately customized. The NCI-60 data set, one of the most reliable and well-known sources of experimentally verified compounds, was used to develop our model. From that data set, we collected and refined data about compounds screened across nine cancer types (panels), including breast, central nervous system, colon, leukemia, nonsmall cell lung, melanoma, ovarian, prostate, and renal, for model training and evaluation. The model performance evaluated on an independent test set shows that iACP-GCR surpasses the three advanced computational methods for multitask learning. The integration of two shortcut connection types in the shared networks also improves the prediction efficiency. We also deployed the model as a public web server to assist the research community in screening potential anticancer compounds.
中文翻译:

图卷积残差神经网络的多任务学习用于筛选多靶点抗癌化合物
最近,已经开发了各种现代实验筛选流程和测定法来寻找有希望的抗癌候选药物。然而,通过实验方法筛选大量化合物的抗癌活性非常耗时且几乎不可行。为了部分解决这个问题,已经提出了一些计算进展。在这项研究中,我们提出了 iACP-GCR,这是一种基于图卷积残差神经网络多任务学习的模型,具有两种类型的快捷连接,用于识别多靶点抗癌化合物。在我们的架构中,图卷积残差神经网络在单独定制之前由所有预测任务共享。 NCI-60 数据集是经过实验验证的化合物最可靠和最知名的来源之一,用于开发我们的模型。从该数据集中,我们收集并完善了在九种癌症类型(面板)中筛选的化合物的数据,包括乳腺癌、中枢神经系统、结肠癌、白血病、非小细胞肺癌、黑色素瘤、卵巢癌、前列腺癌和肾癌,用于模型训练和评估。在独立测试集上评估的模型性能表明,iACP-GCR 超越了三种先进的多任务学习计算方法。共享网络中两种快捷连接类型的集成也提高了预测效率。我们还将该模型部署为公共网络服务器,以协助研究界筛选潜在的抗癌化合物。
更新日期:2024-08-29
中文翻译:

图卷积残差神经网络的多任务学习用于筛选多靶点抗癌化合物
最近,已经开发了各种现代实验筛选流程和测定法来寻找有希望的抗癌候选药物。然而,通过实验方法筛选大量化合物的抗癌活性非常耗时且几乎不可行。为了部分解决这个问题,已经提出了一些计算进展。在这项研究中,我们提出了 iACP-GCR,这是一种基于图卷积残差神经网络多任务学习的模型,具有两种类型的快捷连接,用于识别多靶点抗癌化合物。在我们的架构中,图卷积残差神经网络在单独定制之前由所有预测任务共享。 NCI-60 数据集是经过实验验证的化合物最可靠和最知名的来源之一,用于开发我们的模型。从该数据集中,我们收集并完善了在九种癌症类型(面板)中筛选的化合物的数据,包括乳腺癌、中枢神经系统、结肠癌、白血病、非小细胞肺癌、黑色素瘤、卵巢癌、前列腺癌和肾癌,用于模型训练和评估。在独立测试集上评估的模型性能表明,iACP-GCR 超越了三种先进的多任务学习计算方法。共享网络中两种快捷连接类型的集成也提高了预测效率。我们还将该模型部署为公共网络服务器,以协助研究界筛选潜在的抗癌化合物。

































 京公网安备 11010802027423号
京公网安备 11010802027423号