当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Antibody-SGM, a Score-Based Generative Model for Antibody Heavy-Chain Design
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-27 , DOI: 10.1021/acs.jcim.4c00711 Xuezhi Xie 1, 2 , Pedro A Valiente 1 , Jin Sub Lee 1, 3 , Jisun Kim 1 , Philip M Kim 1, 2, 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-27 , DOI: 10.1021/acs.jcim.4c00711 Xuezhi Xie 1, 2 , Pedro A Valiente 1 , Jin Sub Lee 1, 3 , Jisun Kim 1 , Philip M Kim 1, 2, 3
Affiliation
|
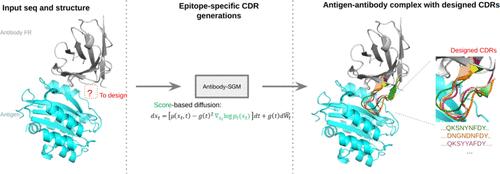
Traditional computational methods for antibody design involved random mutagenesis followed by energy function assessment for candidate selection. Recently, diffusion models have garnered considerable attention as cutting-edge generative models, lauded for their remarkable performance. However, these methods often focus solely on the backbone or sequence, resulting in the incomplete depiction of the overall structure and necessitating additional techniques to predict the missing component. This study presents Antibody-SGM, an innovative joint structure-sequence diffusion model that addresses the limitations of existing protein backbone generation models. Unlike previous models, Antibody-SGM successfully integrates sequence-specific attributes and functional properties into the generation process. Our methodology generates full-atom native-like antibody heavy chains by refining the generation to create valid pairs of sequences and structures, starting with random sequences and structural properties. The versatility of our method is demonstrated through various applications, including the design of full-atom antibodies, antigen-specific CDR design, antibody heavy chains optimization, validation with Alphafold3, and the identification of crucial antibody sequences and structural features. Antibody-SGM also optimizes protein function through active inpainting learning, allowing simultaneous sequence and structure optimization. These improvements demonstrate the promise of our strategy for protein engineering and significantly increase the power of protein design models.
中文翻译:

Antibody-SGM,一种基于评分的抗体重链设计生成模型
抗体设计的传统计算方法涉及随机诱变,然后进行候选选择的能量函数评估。最近,扩散模型作为尖端生成模型受到了相当大的关注,并因其卓越的性能而受到赞誉。然而,这些方法通常只关注主链或序列,导致整体结构的描述不完整,并且需要额外的技术来预测缺失的成分。本研究提出了 Antibody-SGM,这是一种创新的联合结构序列扩散模型,解决了现有蛋白质主链生成模型的局限性。与以前的模型不同,Antibody-SGM 成功地将序列特异性属性和功能特性集成到生成过程中。我们的方法通过从随机序列和结构特性开始改进生成以创建有效的序列和结构对来生成全原子天然样抗体重链。我们的方法的多功能性通过各种应用得到证明,包括全原子抗体的设计、抗原特异性 CDR 设计、抗体重链优化、Alphafold3 验证以及关键抗体序列和结构特征的鉴定。 Antibody-SGM 还通过主动修复学习来优化蛋白质功能,从而实现同步序列和结构优化。这些改进证明了我们蛋白质工程策略的前景,并显着增强了蛋白质设计模型的能力。
更新日期:2024-08-29
中文翻译:

Antibody-SGM,一种基于评分的抗体重链设计生成模型
抗体设计的传统计算方法涉及随机诱变,然后进行候选选择的能量函数评估。最近,扩散模型作为尖端生成模型受到了相当大的关注,并因其卓越的性能而受到赞誉。然而,这些方法通常只关注主链或序列,导致整体结构的描述不完整,并且需要额外的技术来预测缺失的成分。本研究提出了 Antibody-SGM,这是一种创新的联合结构序列扩散模型,解决了现有蛋白质主链生成模型的局限性。与以前的模型不同,Antibody-SGM 成功地将序列特异性属性和功能特性集成到生成过程中。我们的方法通过从随机序列和结构特性开始改进生成以创建有效的序列和结构对来生成全原子天然样抗体重链。我们的方法的多功能性通过各种应用得到证明,包括全原子抗体的设计、抗原特异性 CDR 设计、抗体重链优化、Alphafold3 验证以及关键抗体序列和结构特征的鉴定。 Antibody-SGM 还通过主动修复学习来优化蛋白质功能,从而实现同步序列和结构优化。这些改进证明了我们蛋白质工程策略的前景,并显着增强了蛋白质设计模型的能力。































 京公网安备 11010802027423号
京公网安备 11010802027423号