当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Predicting the Dynamic Interaction of Intrinsically Disordered Proteins
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-20 , DOI: 10.1021/acs.jcim.4c00930 Yuchuan Zheng 1 , Qixiu Li 1 , Maria I Freiberger 2 , Haoyu Song 1 , Guorong Hu 1 , Moxin Zhang 1 , Ruoxu Gu 3 , Jingyuan Li 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-20 , DOI: 10.1021/acs.jcim.4c00930 Yuchuan Zheng 1 , Qixiu Li 1 , Maria I Freiberger 2 , Haoyu Song 1 , Guorong Hu 1 , Moxin Zhang 1 , Ruoxu Gu 3 , Jingyuan Li 1
Affiliation
|
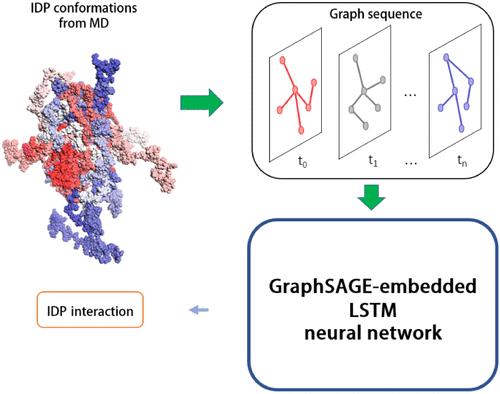
Intrinsically disordered proteins (IDPs) participate in various biological processes. Interactions involving IDPs are usually dynamic and are affected by their inherent conformation fluctuations. Comprehensive characterization of these interactions based on current techniques is challenging. Here, we present GSALIDP, a GraphSAGE-embedded LSTM network, to capture the dynamic nature of IDP-involved interactions and predict their behaviors. This framework models multiple conformations of IDP as a dynamic graph, which can effectively describe the fluctuation of its flexible conformation. The dynamic interaction between IDPs is studied, and the data sets of IDP conformations and their interactions are obtained through atomistic molecular dynamic (MD) simulations. Residues of IDP are encoded through a series of features including their frustration. GSALIDP can effectively predict the interaction sites of IDP and the contact residue pairs between IDPs. Its performance in predicting IDP interactions is on par with or even better than the conventional models in predicting the interaction of structural proteins. To the best of our knowledge, this is the first model to extend the protein interaction prediction to IDP-involved interactions.
中文翻译:

预测本质无序蛋白质的动态相互作用
本质无序蛋白 (IDP) 参与各种生物过程。涉及 IDP 的相互作用通常是动态的,并受到其固有构象波动的影响。基于当前技术对这些相互作用进行全面表征具有挑战性。在这里,我们提出了 GSALIDP,一种 GraphSAGE 嵌入的 LSTM 网络,用于捕获 IDP 涉及的交互的动态性质并预测其行为。该框架将IDP的多种构象建模为动态图,可以有效地描述其灵活构象的波动。研究IDP之间的动态相互作用,通过原子分子动力学(MD)模拟获得IDP构象及其相互作用的数据集。 IDP 的残留物通过一系列特征(包括其挫败感)进行编码。 GSALIDP可以有效预测IDP的相互作用位点以及IDP之间的接触残基对。它在预测 IDP 相互作用方面的性能与预测结构蛋白相互作用的传统模型相当甚至更好。据我们所知,这是第一个将蛋白质相互作用预测扩展到 IDP 涉及的相互作用的模型。
更新日期:2024-08-20
中文翻译:

预测本质无序蛋白质的动态相互作用
本质无序蛋白 (IDP) 参与各种生物过程。涉及 IDP 的相互作用通常是动态的,并受到其固有构象波动的影响。基于当前技术对这些相互作用进行全面表征具有挑战性。在这里,我们提出了 GSALIDP,一种 GraphSAGE 嵌入的 LSTM 网络,用于捕获 IDP 涉及的交互的动态性质并预测其行为。该框架将IDP的多种构象建模为动态图,可以有效地描述其灵活构象的波动。研究IDP之间的动态相互作用,通过原子分子动力学(MD)模拟获得IDP构象及其相互作用的数据集。 IDP 的残留物通过一系列特征(包括其挫败感)进行编码。 GSALIDP可以有效预测IDP的相互作用位点以及IDP之间的接触残基对。它在预测 IDP 相互作用方面的性能与预测结构蛋白相互作用的传统模型相当甚至更好。据我们所知,这是第一个将蛋白质相互作用预测扩展到 IDP 涉及的相互作用的模型。































 京公网安备 11010802027423号
京公网安备 11010802027423号