当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Reinforcing Tunnel Network Exploration in Proteins Using Gaussian Accelerated Molecular Dynamics
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-15 , DOI: 10.1021/acs.jcim.4c00966
Nishita Mandal 1, 2 , Bartlomiej Surpeta 1, 2 , Jan Brezovsky 1, 2
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-08-15 , DOI: 10.1021/acs.jcim.4c00966
Nishita Mandal 1, 2 , Bartlomiej Surpeta 1, 2 , Jan Brezovsky 1, 2
Affiliation
|
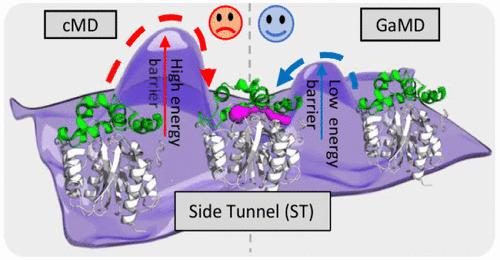
Tunnels are structural conduits in biomolecules responsible for transporting chemical compounds and solvent molecules from the active site. They have been shown to be present in a wide variety of enzymes across all functional and structural classes. However, the study of such pathways is experimentally challenging, because they are typically transient. Computational methods, such as molecular dynamics (MD) simulations, have been successfully proposed to explore tunnels. Conventional MD (cMD) provides structural details to characterize tunnels but suffers from sampling limitations to capture rare tunnel openings on longer time scales. Therefore, in this study, we explored the potential of Gaussian accelerated MD (GaMD) simulations to improve the exploration of complex tunnel networks in enzymes. We used the haloalkane dehalogenase LinB and its two variants with engineered transport pathways, which are not only well-known for their application potential but have also been extensively studied experimentally and computationally regarding their tunnel networks and their importance in multistep catalytic reactions. Our study demonstrates that GaMD efficiently improves tunnel sampling and allows the identification of all known tunnels for LinB and its two mutants. Furthermore, the improved sampling provided insight into a previously unknown transient side tunnel (ST). The extensive conformational landscape explored by GaMD simulations allowed us to investigate in detail the mechanism of ST opening. We determined variant-specific dynamic properties of ST opening, which were previously inaccessible due to limited sampling of cMD. Our comprehensive analysis supports multiple indicators of the functional relevance of the ST, emphasizing its potential significance beyond structural considerations. In conclusion, our research proves that the GaMD method can overcome the sampling limitations of cMD for the effective study of tunnels in enzymes, providing further means for identifying rare tunnels in enzymes with the potential for drug development, precision medicine, and rational protein engineering.
中文翻译:

使用高斯加速分子动力学加强蛋白质中的隧道网络探索
隧道是生物分子中的结构管道,负责从活性位点运输化合物和溶剂分子。它们已被证明存在于所有功能和结构类别的多种酶中。然而,对此类途径的研究在实验上具有挑战性,因为它们通常是短暂的。分子动力学(MD)模拟等计算方法已被成功提出来探索隧道。传统 MD (cMD) 提供结构细节来表征隧道,但在较长时间尺度上捕获罕见的隧道开口时受到采样限制。因此,在这项研究中,我们探索了高斯加速 MD (GaMD) 模拟在改进酶中复杂隧道网络探索方面的潜力。我们使用卤代烷脱卤酶 LinB 及其两种具有工程化转运途径的变体,它们不仅因其应用潜力而闻名,而且还对其隧道网络及其在多步催化反应中的重要性进行了广泛的实验和计算研究。我们的研究表明,GaMD 有效地改进了隧道采样,并允许识别 LinB 及其两个突变体的所有已知隧道。此外,改进的采样可以深入了解以前未知的瞬态侧隧道 (ST)。 GaMD 模拟探索的广泛构象景观使我们能够详细研究 ST 打开的机制。我们确定了 ST 打开的变体特异性动态特性,这在以前由于 cMD 采样有限而无法获得。 我们的综合分析支持 ST 功能相关性的多个指标,强调其超越结构考虑的潜在意义。总之,我们的研究证明,GaMD 方法可以克服 cMD 的采样限制,有效研究酶中的通道,为识别酶中的稀有通道提供进一步的手段,具有药物开发、精准医学和合理蛋白质工程的潜力。
更新日期:2024-08-15
中文翻译:

使用高斯加速分子动力学加强蛋白质中的隧道网络探索
隧道是生物分子中的结构管道,负责从活性位点运输化合物和溶剂分子。它们已被证明存在于所有功能和结构类别的多种酶中。然而,对此类途径的研究在实验上具有挑战性,因为它们通常是短暂的。分子动力学(MD)模拟等计算方法已被成功提出来探索隧道。传统 MD (cMD) 提供结构细节来表征隧道,但在较长时间尺度上捕获罕见的隧道开口时受到采样限制。因此,在这项研究中,我们探索了高斯加速 MD (GaMD) 模拟在改进酶中复杂隧道网络探索方面的潜力。我们使用卤代烷脱卤酶 LinB 及其两种具有工程化转运途径的变体,它们不仅因其应用潜力而闻名,而且还对其隧道网络及其在多步催化反应中的重要性进行了广泛的实验和计算研究。我们的研究表明,GaMD 有效地改进了隧道采样,并允许识别 LinB 及其两个突变体的所有已知隧道。此外,改进的采样可以深入了解以前未知的瞬态侧隧道 (ST)。 GaMD 模拟探索的广泛构象景观使我们能够详细研究 ST 打开的机制。我们确定了 ST 打开的变体特异性动态特性,这在以前由于 cMD 采样有限而无法获得。 我们的综合分析支持 ST 功能相关性的多个指标,强调其超越结构考虑的潜在意义。总之,我们的研究证明,GaMD 方法可以克服 cMD 的采样限制,有效研究酶中的通道,为识别酶中的稀有通道提供进一步的手段,具有药物开发、精准医学和合理蛋白质工程的潜力。







































 京公网安备 11010802027423号
京公网安备 11010802027423号