当前位置:
X-MOL 学术
›
Ecol. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
BioEncoder: A metric learning toolkit for comparative organismal biology
Ecology Letters ( IF 7.6 ) Pub Date : 2024-08-13 , DOI: 10.1111/ele.14495 Moritz D Lürig 1, 2 , Emanuela Di Martino 3, 4 , Arthur Porto 1, 5
Ecology Letters ( IF 7.6 ) Pub Date : 2024-08-13 , DOI: 10.1111/ele.14495 Moritz D Lürig 1, 2 , Emanuela Di Martino 3, 4 , Arthur Porto 1, 5
Affiliation
|
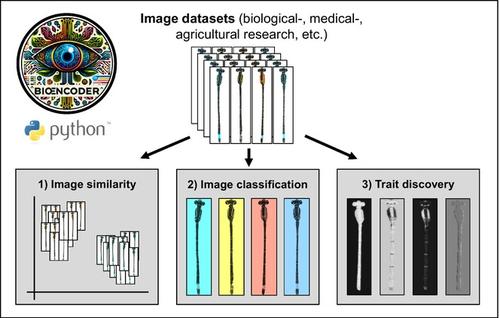
In the realm of biological image analysis, deep learning (DL) has become a core toolkit, for example for segmentation and classification. However, conventional DL methods are challenged by large biodiversity datasets characterized by unbalanced classes and hard‐to‐distinguish phenotypic differences between them. Here we present BioEncoder, a user‐friendly toolkit for metric learning, which overcomes these challenges by focussing on learning relationships between individual data points rather than on the separability of classes. BioEncoder is released as a Python package, created for ease of use and flexibility across diverse datasets. It features taxon‐agnostic data loaders, custom augmentation options, and simple hyperparameter adjustments through text‐based configuration files. The toolkit's significance lies in its potential to unlock new research avenues in biological image analysis while democratizing access to advanced deep metric learning techniques. BioEncoder focuses on the urgent need for toolkits bridging the gap between complex DL pipelines and practical applications in biological research.
中文翻译:

BioEncoder:比较有机体生物学的度量学习工具包
在生物图像分析领域,深度学习(DL)已成为核心工具包,例如用于分割和分类。然而,传统的深度学习方法面临着大型生物多样性数据集的挑战,这些数据集的特征是类别不平衡和难以区分它们之间的表型差异。在这里,我们提出了 BioEncoder,一个用户友好的度量学习工具包,它通过专注于学习各个数据点之间的关系而不是类的可分离性来克服这些挑战。 BioEncoder 以 Python 包的形式发布,旨在实现跨不同数据集的易用性和灵活性。它具有与分类无关的数据加载器、自定义增强选项以及通过基于文本的配置文件进行简单的超参数调整。该工具包的重要性在于它有可能开启生物图像分析的新研究途径,同时使先进的深度度量学习技术的访问民主化。 BioEncoder 重点关注对弥合复杂深度学习管道与生物研究实际应用之间差距的工具包的迫切需求。
更新日期:2024-08-13
中文翻译:

BioEncoder:比较有机体生物学的度量学习工具包
在生物图像分析领域,深度学习(DL)已成为核心工具包,例如用于分割和分类。然而,传统的深度学习方法面临着大型生物多样性数据集的挑战,这些数据集的特征是类别不平衡和难以区分它们之间的表型差异。在这里,我们提出了 BioEncoder,一个用户友好的度量学习工具包,它通过专注于学习各个数据点之间的关系而不是类的可分离性来克服这些挑战。 BioEncoder 以 Python 包的形式发布,旨在实现跨不同数据集的易用性和灵活性。它具有与分类无关的数据加载器、自定义增强选项以及通过基于文本的配置文件进行简单的超参数调整。该工具包的重要性在于它有可能开启生物图像分析的新研究途径,同时使先进的深度度量学习技术的访问民主化。 BioEncoder 重点关注对弥合复杂深度学习管道与生物研究实际应用之间差距的工具包的迫切需求。






























 京公网安备 11010802027423号
京公网安备 11010802027423号